Figure 1.
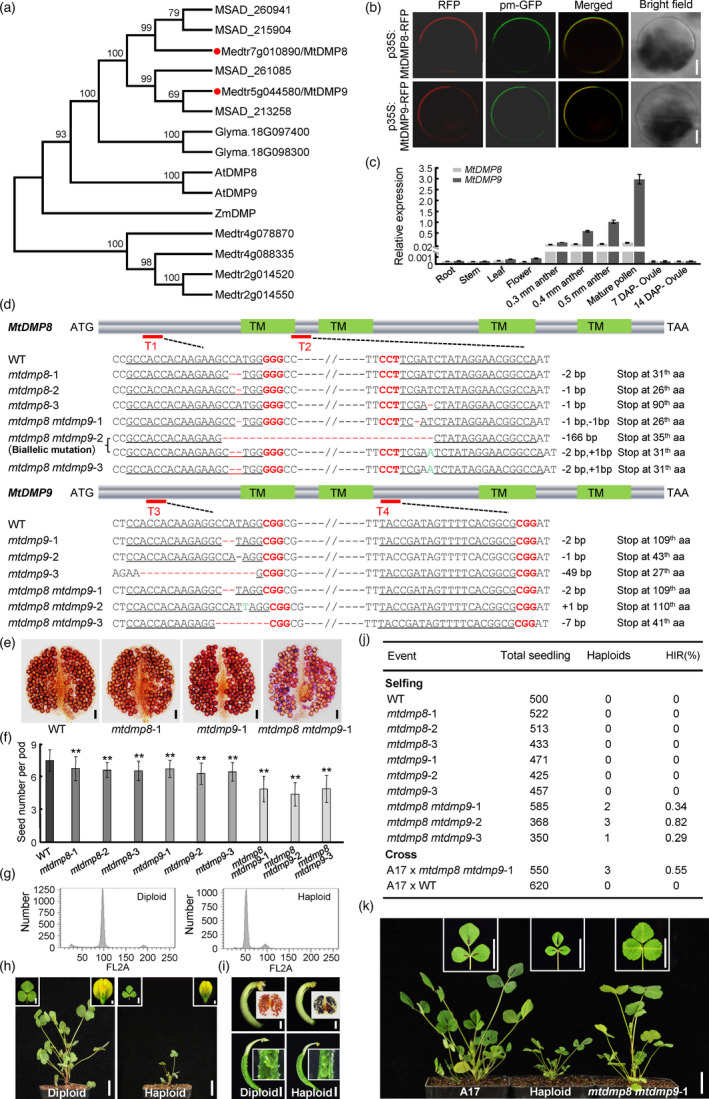
Inactivation of DMP homologues triggers haploid induction in Medicago truncatula. (a) Phylogenetic analysis of ZmDMP and its homologues in M. truncatula (Medtr or Mt), Arabidopsis (At), alfalfa (MSAD) and soybean (Glyma.). MtDMP8 and MtDMP9 are highlighted with red dots. Full‐length protein sequences were aligned using ClustalW, and a neighbour‐joining phylogenetic tree was constructed using MEGA6 software. Numbers on branches indicate bootstrap percentages for 1000 replicates. (b) Subcellular localization of MtDMP8‐RFP and MtDMP9‐RFP proteins in Arabidopsis leaf protoplasts; pm‐GFP was used as a plasma membrane marker. Bars, 5 μm. (c) Relative transcript levels of MtDMP8 and MtDMP9 in the indicated tissues, as determined by RT‐qPCR. MtActin was used as an internal control. Values are means ± SD of three technical replicates. Three independent experiments were performed, with similar results. (d) Schematic representation of MtDMP8 and MtDMP9 gene structures and genome editing experimental design. Filled blocks indicate the coding region. Green blocks correspond to the regions encoding the four predicted transmembrane domains (TMs). Red lines indicate the four regions (T1–4) targeted by sgRNAs. The relevant sequences from the wild‐type (WT) and mutant alleles are shown below the gene structure schematics. (e) Pollen viability assays with Alexander’s stain in the T1 progeny of selfed WT and mtdmp mutants. Bars, 50 μm. (f) Comparison of seed number per pod in the T1 progeny of selfed WT and mtdmp mutants. Bars represent means ± SD (n = 30); asterisks indicate significant differences from the WT (**P < 0.01, Student’s t‐test). (g) Confirmation of ploidy by flow cytometry analysis. (h) Phenotypic differences between M. truncatula haploid and diploid plants (whole plant, leaf and flower). Bars, 2 cm for whole plant; 5 mm for leaf; and 1 mm for flower. (i) Comparison of anther and pollen viability, as well as carpels and ovules between haploid and diploid M. truncatula plants. Bars, 1 mm. (j) Haploid induction rate (HIR) determined by self‐pollination or crossing. For crossing, the M. truncatula ecotype A17 was used as the female parent and was pollinated with mtdmp8 mtdmp9‐1. (k) Representative haploid plant from crossing. Bars, 2 cm.
