Fig. 2.
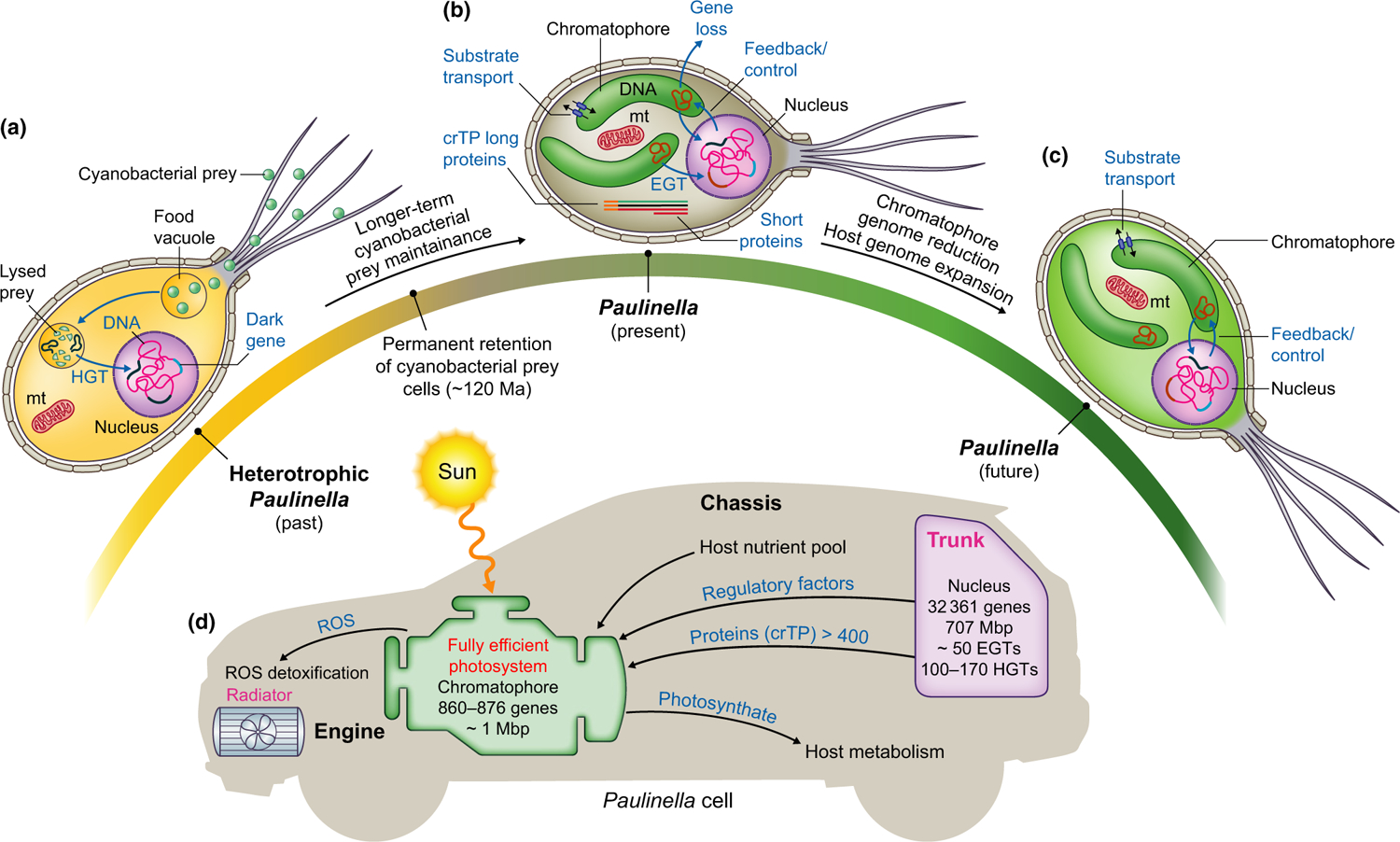
Putative timeline of plastid evolution in Paulinella encompassing both a historical and future perspective based on the chassis and engine model. (a) Heterotrophic Paulinella ingesting cyanobacterial cells likely using filose, feeding pseudopodia as a result of the presence of silica scales on the cell surface (Johnson et al., 1988). DNA from ingested bacterial cells can enter the nucleus and become integrated into the host genome via horizontal gene transfer (HGT). These foreign genes, as well as other novel ‘dark’ genes that evolved in this lineage, enabled the host to maintain the cyanobacteria in food vacuoles for increasingly longer periods of time until the association became permanent. (b) The status of extant photosynthetic Paulinella showing the two chromatophores and current evolutionary processes. (c) Putative future status of photosynthetic Paulinella. The chromatophore will encode only essential genes that cannot be transferred to the host genome because the encoded proteins either are problematic to transport into the chromatophore or require redox regulationwithin the organelle(e.g. co-locationfor redox regulation (CoRR hypothesis); Allen, 2017).The chromatophore may also lose its peptidoglycan layer, as observed in other lineages containing primary plastids (except Glaucophyta). Adaptations will probably increase the Paulinella growth rate and the ability to deal with high light, expanding its environmental niche. (d) The chassis and engine model of plastid endosymbiosis. The approximate size of the chromatophore genome and the number of genes it encodes across Paulinella species are shown inside the engine. Parts of the model where questions remain about the endosymbiosis in Paulinella are shown in red text (for more detail see Box 1). crTP, chromatophore targeting peptide; EGT, endosymbiotic gene transfer; mt, mitochondrion; ROS, reactive oxygen species.
