Extended Data Figure 9 |. MIPS521 stabilises the A1R-Gi2 ternary complex.
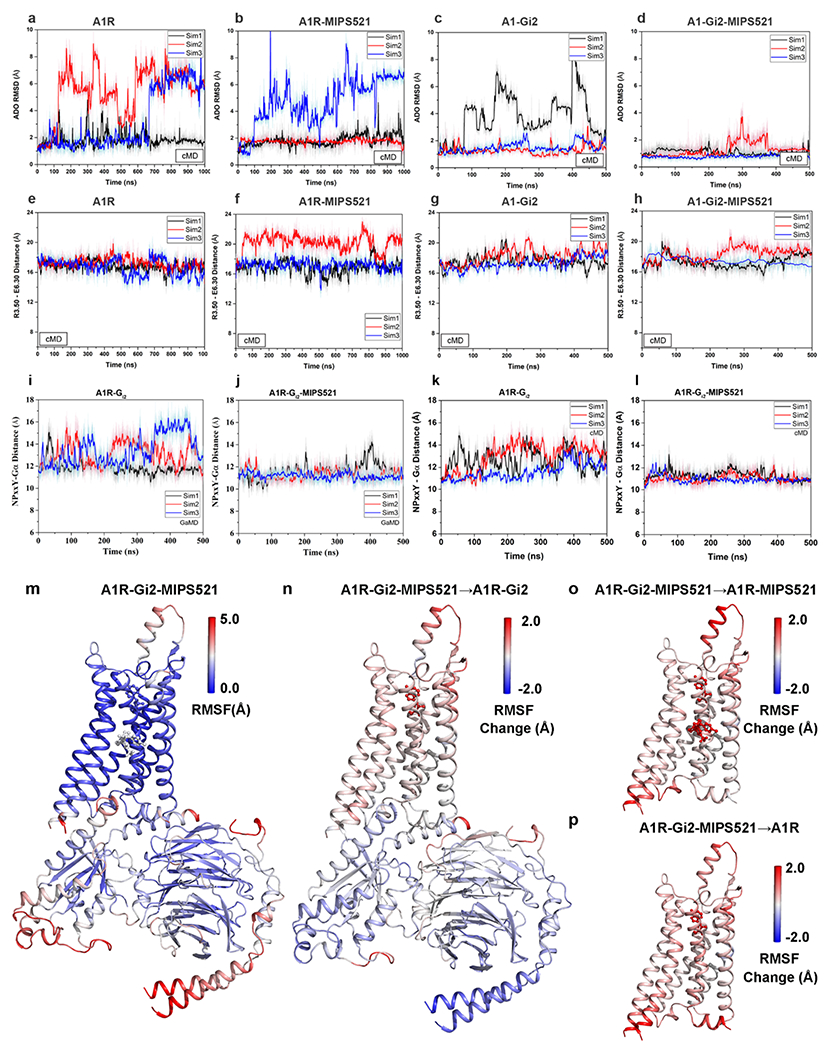
a-d, RMSD (Å) of ADO from cMD simulations completed in the (a) absence or (b) presence of MIPS521, (c) Gi2, or (d) both Gi2 and MIPS521. e-h, Distance between the intracellular ends of TM3 and TM6 (measured as the distance in Å between Arg1053.50 and Glu2296.30) in the (e) absence or (f) presence of MIPS521, (g) Gi2, or (h) both Gi2 and MIPS521. Each condition represents three cMD simulations, with each simulation trace displayed in a different colour (black, red, blue). The lines depict the running average over 2 ns. i,j, Distance between A1R and Gi2 (measured as the distance in Å between the NPxxY motif of A1R and the C terminus of the Gα α5 helix) from GaMD simulations in the (i) absence and (j) presence of MIPS521. k,l, Distance between A1R and Gi2 from cMD simulations in the (k) absence and (l) presence of MIPS521. Each condition represents three GaMD/cMD simulations, with each simulation trace displayed in a different colour (black, red, blue). Thick lines depict the running average over 2 ns. m-p, Flexibility change upon removal of PAM and/or Gi2 protein from the ADO-bound A1R obtained from GaMD simulations. (m) Root-mean-square fluctuations (RMSFs) of the A1R-Gi2-MIPS521. A colour scale of 0.0 Å (blue) to 5.0 Å (red) was used. (n) Change in the RMSFs of the A1R-Gi2 when MIPS521 was removed from A1R-Gi2-MIPS521. (o) Change in the RMSFs of the A1R and MIPS521 when the Gi2 was removed from A1R-Gi2-MIPS521. (p) Change in the RMSFs of the A1R when the Gi2 and MIPS521 were removed from A1R-Gi2-MIPS521 system. A colour scale of −2.0 Å (blue) to 2.0 Å (red) was used for n, o and p.
