Extended Data Figure 4 |. Cryo-EM data processing for the MIPS521-ADO-A1R-Gi2 (a) and ADO-A1R-Gi2 (b) complexes.
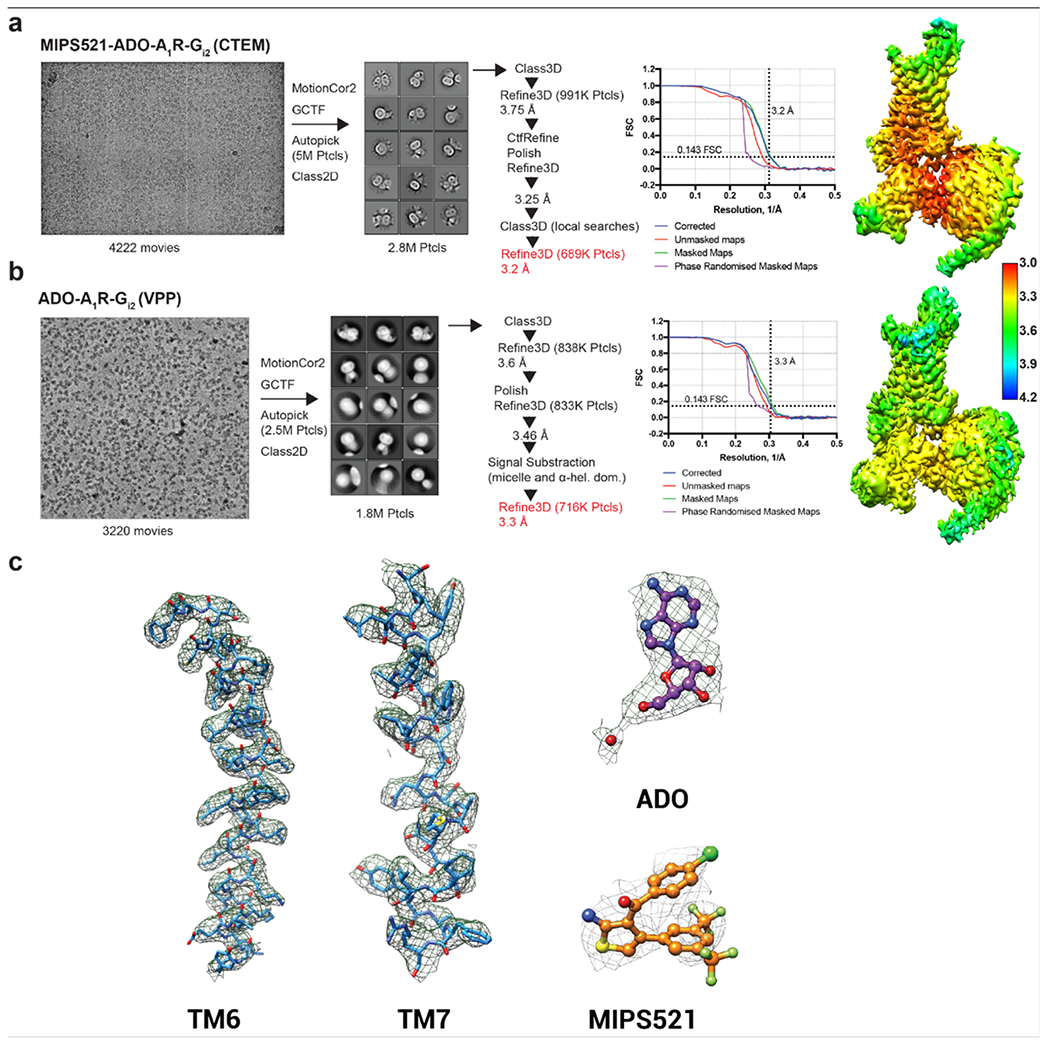
Representative cryo-EM micrographs of each of the complexes. Reference-free 2D class averages of the complexes in LMNG and CHS detergent micelles. Gold-standard Fourier shell correlation (FSC) curves, showing the overall nominal resolution of 3.2 Å and 3.3 Å, respectively, at FSC 0.143. Corresponding 3D cryo-EM maps coloured according to local resolution estimation (Å) in Relion. c, Atomic resolution model of representative regions from the MIPS521-ADO-A1R-Gi2 structure of the A1R transmembrane domain, ADO, and MIPS521. The molecular model is shown in ball and stick representation, coloured by heteroatom, and the cryo-EM map displayed in mesh contoured at 0.02.
