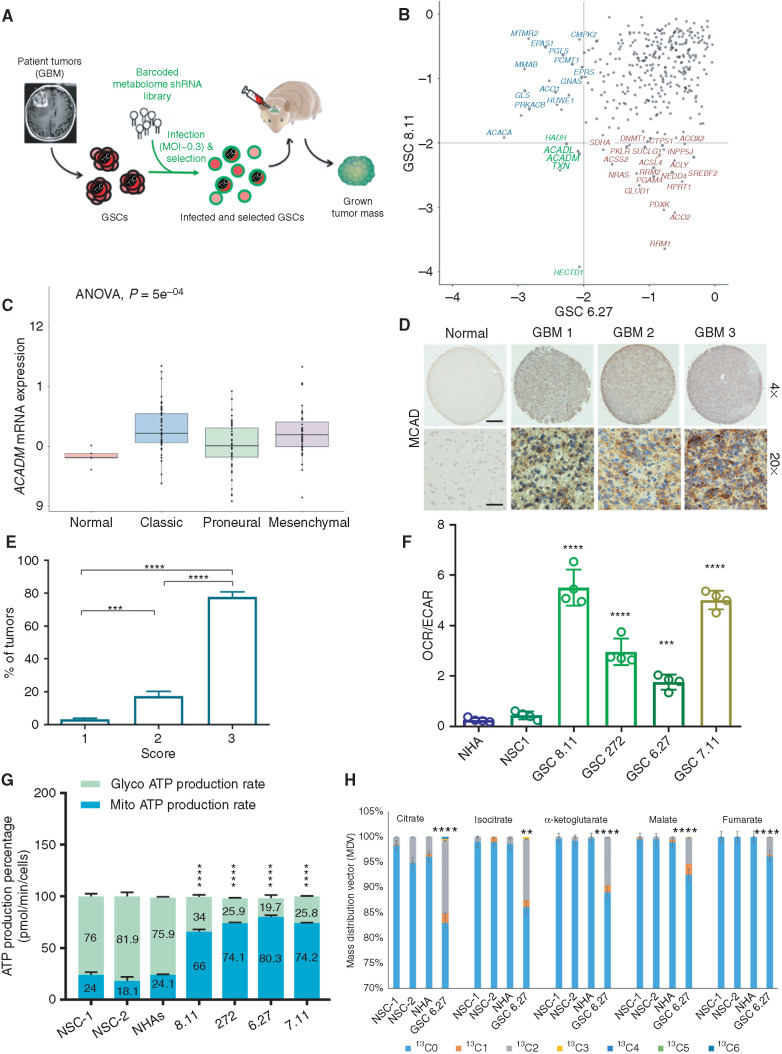
Figure 1.
ACADM emerges as a clinically relevant dependency of GBM. A, Schematics of experimental design for intracranial metabolome shRNA screens in patient-derived GSCs. The lentiviral library was transduced at a low MOI (less than one integrant/cell). B, Gene–rank analysis highlighting the behavior of genes involved in FA metabolism (ACADM, ACADL, PRKACB, and ACSL6) in in vivo screens executed in two independent GSC models: GSC 8.11 and GSC 6.27 (RSA, logP). C,ACADM mRNA levels in glioma subtypes vs. normal brain (TCGA data set). Log2 RSEM values were calculated to compare expression levels of ACADM and other genes involved in the FA synthesis pathway. Using the GBM data set (n = 167), analysis was performed comparing expression among normal tissue (n = 5), classic (n = 68), proneural (n = 46), and mesenchymal (n = 48) subtypes. ANOVA was performed across all groups to determine statistical significance, with pairwise Wilcoxon tests to identify which subtypes showed a difference. D, Immunohistochemistry for MCAD on tissue microarray (TMA) derived from normal brain and GBM tissue. Scale bars, 100 μm for ×4 and 25 μm for ×20. E, GBM percentage distribution based on MCAD expression levels in three independent TMAs. The scores 1 to 3 were independently determined using the following scoring system to approximate the percentage of cells positive for staining with the MCAD antibody: 1 = 0% to 10%, 2 = 11% to 50%, and 3 = 51% to 100%. Representative tissue scoring is presented in D. Data represent the analysis of three independent TMAs. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. ***, P = 0.0002; ****, P < 0.0001. F, Bioenergetic profiling of NHA, NSC, and GSC lines using Seahorse technology. Basal oxygen consumption rate (OCR; pMoles/minute) and extracellular acidification rate (ECAR; mpH/minute) were used for calculations. Values represent the mean ± SD of four independent experiments. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. ***, P = 0.0003; ****, P = 0.0001. G, Quantification of energy production in the indicated cell lines by Seahorse XF Real-Time ATP Rate Assay. MitoATP Production Rate and glycoATP Production Rate were calculated from OCR and ECAR measurements under basal conditions. Values are expressed as mean ± SD; P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. ****, P < 0.0001. H, Isotopolog patterns for incorporation of 13C-labeled oleate into TCA cycle intermediates, as measured by LC/MS in NHAs, NSCs, and GSCs in basal conditions. Cells were cultured with 13C-oleate for 6 hours prior to sample collection. N = 4 biological replicates, error = ±SD. **, P = 0.0014; ****, P < 0.0001.