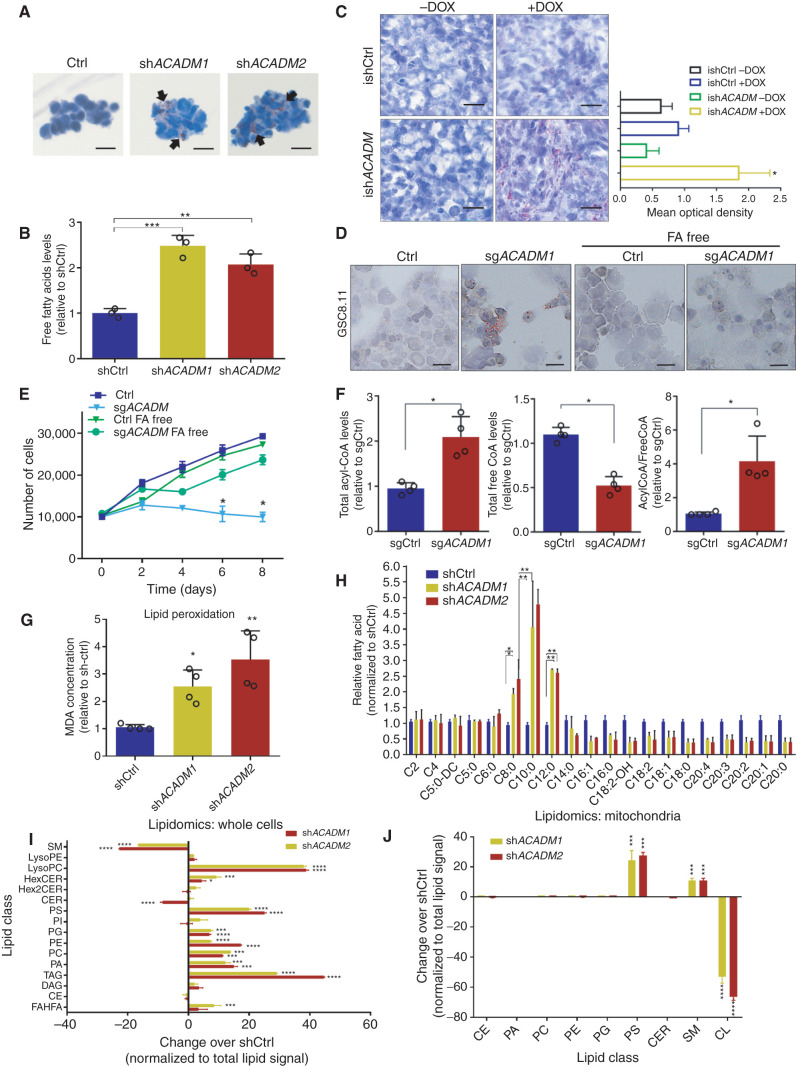
Figure 4.
Accumulation of MCFAs induces toxic alterations of lipid metabolism in GSCs. A, Oil Red O staining in GSC 8.11 cells infected with anti-ACADM or nontargeting shRNA in vitro. Black arrows indicate sites of lipid accumulation. Scale bar, 20 μm. B, Colorimetric determination of free FAs from GSC whole-cell extracts. Values represent the mean ± SD of three independent experiments. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. **, P = 0.0061; ***, P = 0.0003. C, Oil Red O staining in xenograft tumor tissues derived from GSC 8.11 infected with inducible shRNA (ishACADM) constructs. Doxycycline was administered approximately 20 days after cell implantation. Oil Red O staining quantification of tumor tissues shown in C was obtained using ImageJ software analysis. Scale bar, 25 μm. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P = 0.033. d and E, Oil Red O staining (D) and growth curve (E) of ACADM wild-type or null GSC 8.11 grown in normal or FA-free medium. Cells were selected with puromycin for 48 hours prior to starting the experiment. Oil Red O staining in cells after 48-hour puromycin selection. Scale bar in D, 10 μm. Data in E represent the mean ± SD of three biologically independent replicates. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. **, P ≤ 0.007. F, Colorimetric analysis of acyl-CoA species in MCAD-deficient GSC 8.11 cells. Data represent the mean ± SEM of four biologically independent replicates. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P < 0.03. G, Quantification of lipid peroxidation determined by measuring the production of malondialdehyde (MDA) using the Colorimetric Microplate Assay for Lipid Peroxidation Kit. Data represent the mean ± SD of four biologically independent replicates. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P = 0.02; **, P = 0.01. H, Quantitative LC/MS-MS lipid profiling of FFA content of ACADM wild-type or null GSC 8.11 cells. Data represent mean ± SEM of three biologically independent replicates. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P < 0.03; **, P < 0.004. i and J, Relative amount of total lipid classes measured by mass spectrometry of whole-cell extracts (I) or mitochondria (J) from GSC 8.11 cells infected with shRNA targeting ACADM. Data are reported as fold change over control cells infected with nontargeting shRNA. Mean values ± SD of three biologically independent replicates. P values were generated using Kruskal–Wallis ANOVA. Dunn test for comparison among groups. *, P < 0.05; ***, P = 0.001; ****, P = 0.0001. CE, cholesteryl ester; CER, ceramide; DAG, diacylglycerol; FAHFA, fatty acid ester of hydroxyl fatty acids; Hex2CER, dihexosylceramide; HexCER, hexosylceramide; LPC, lyso-phosphatidylcholine; LPE, lyso-phosphatidylethanolamine; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine; SM, sphingomyelin; TAG, triacylglycerol.