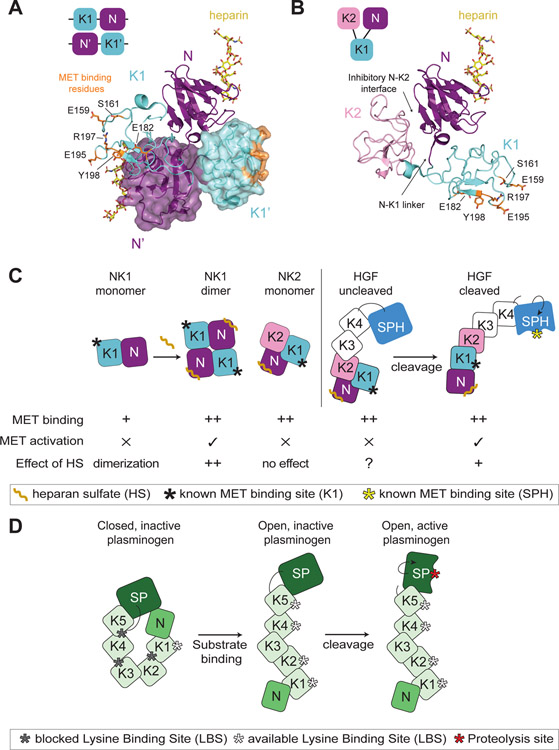
Figure 4: Insights into HGF autoinhibition from the structures of NK1 and NK2 ligands.
(A) Domain organization of the head-to-tail dimer of NK1 is shown schematically in the top left corner. Crystal structure of the NK1 dimer is shown in cartoon representation, with the second monomer shown in a semi-transparent surface mode (PDB: 1GMO). Heparan sulfate is shown in yellow bound to each N domain. Residues important for MET binding in the K1 domain are shown in orange in stick representation. The linker region between the N and K1 domains as well as reciprocal interfaces of the N and K1’ and K1 and N’ domains are shown to contribute to the dimer interface. (B) Domain organization of the autoinhibited NK2 monomer is shown schematically in the top left corner. The NK2 crystal structure is shown in cartoon representation (PDB: 3SP8). The heparan sulfate is modelled in via the alignment of the N domain of the NK2 ligand on the N domain of the NK1 ligand (PDB: 1GMO). The obscured N-K1 linker is indicated by an arrow. (C) Cartoon schematic of NK1, NK2 and HGF (cleaved and uncleaved). The relative binding affinity of these different ligand states for MET is indicated by ‘+’ (low affinity) or ‘++’ (high affinity). The ability of these ligand states to activate MET signaling is denoted by ‘✕’ or ‘✓’. Heparan sulfate (HS) binding to the N domain has different effects on these ligands. NK1: increased binding affinity for MET and ligand dimerization (++), NK2: no effect, HGF-uncleaved: no defined contribution (?), HGF-cleaved: HS promotes HGF dimerization (+) but is not required for signaling. The binding site for MET in the K1 domain and the cleaved SPH domain is indicated by a black and yellow star, respectively. (D) Cartoon schematic showing domain architecture and activation of plasminogen. The inactive molecule is maintained via multiple autoinhibitory intramolecular interactions. Substrate binding by the exposed LBS of K1 domain leads to elongation and accessibility to the additional LBS sites and of the cleavage motif. Maturation of plasminogen by cleavage activates the serine protease domain (SP).