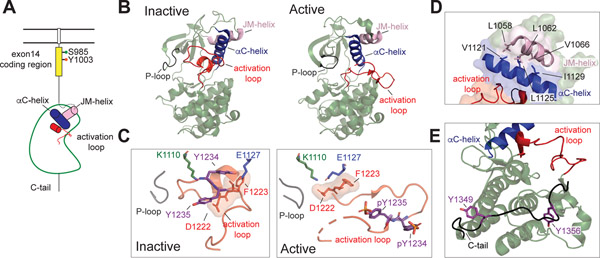
Figure 5: MET kinase domain states.
(A) Cartoon representation of the MET receptor intracellular domains. The region encoded by exon 14 is colored yellow. Two residues (Ser 985, Tyr 1003) that are important for MET regulation upon being phosphorylated are marked. The αC helix and JM helix in the kinase domain are indicated in blue and pink, respectively. The activation loop is shown in red. (B) Crystal structures of an inactive (PDB: 2G15) and active (PDB: 3R7O) MET kinase domain are shown in cartoon representation. (C, left panel) The activation loop of the inactive MET structure forms a short helix that obscures the αC-helix from swinging into the active site and from forming the salt bridge required for catalysis (Lys 1110 and Glu 1127). In this Src/CDK-like inactive conformation, the DFG motif is in the DFG-in conformation, as indicated by the Asp 1222 pointing towards the active site. The JM-helix is shown in light pink. (C, right panel) In the active conformation, the activation loop tyrosines (Tyr 1234 and Tyr 1235) are phosphorylated (denoted as ‘pY’), releasing the activation loop from the active site. (D) Zoomed-in view of the hydrophobic residues that form the interface between the JM helix and the αC helix in the MET kinase domain (PDB: 2G15), including Leu 1058, Leu 1062, Val 1066 on the JM-helix and Val 1121, Leu 1125, Iso 1129 on the αC-helix. (E) The kinase proximal region of the C-terminal tail (Gly 1346-Lys 1360) is shown in black bound to the back of the kinase C-lobe (PDB: 1R0P). Tyr 1349 and Tyr 1356 are shown in purple.