Fig 6. Adaptability to biopsy slides.
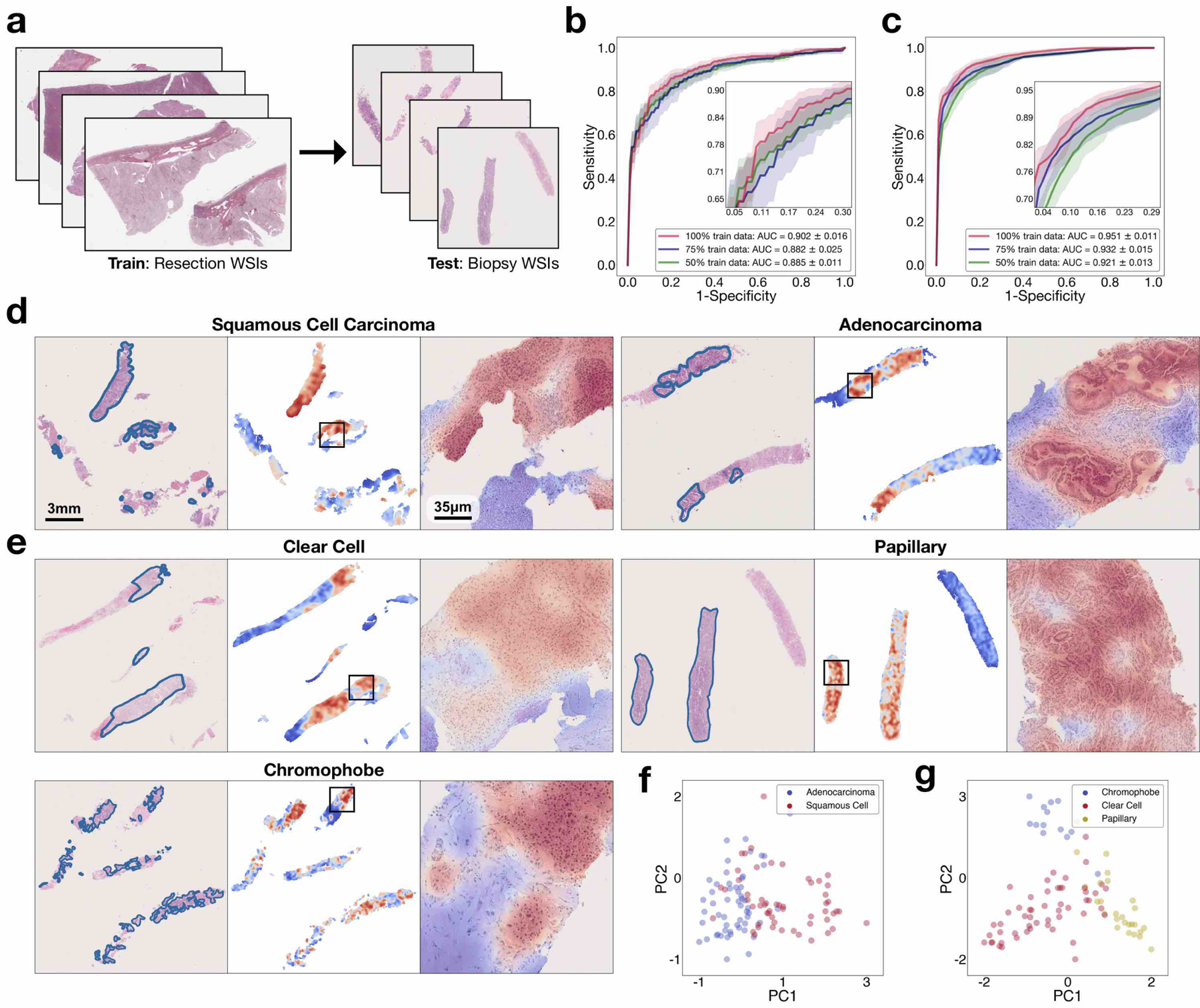
a, Compared with resection WSIs, biopsy WSIs generally contain a much lower tissue content (for example, the average number of patches extracted from the tissue regions of each slide is 820 in our BWH lung biopsy dataset compared with 24,714 in the lung-resection dataset). The presence of crush artefacts as well as poorly differentiated and sparsely distributed tumour cells can further challenge accurate diagnosis. b,c, We observed that CLAM models trained on resections are directly adaptable to biopsy WSIs, achieving a respectable average test AUC of 0.902 ± 0.016 and 0.951 ± 0.011 on our NSCLC (b; n = 110) and RCC (c; n = 92) biopsy independent test cohorts, respectively, without further fine-tuning or ROI extraction. Insets: zoomed-in view of the curves. d,e, Attention heatmap visualization for NSCLC (d) and RCC (e) biopsy slides. H&E slide with annotation by the pathologist for tumour regions (left). Heatmap for patches tiled with a 95% overlap (middle). Zoomed-in view of tumour regions attended by the CLAM model (right). Consistent with our findings on the resection and smartphone datasets, the regions that were most strongly attended by the model consistently correspond to tumour tissue. The attention heatmaps also tend to clearly highlight the tumour–normal tissue boundaries, despite the fact that no patch-level or pixel-level annotation was required or used during training. f,g, The slide-level feature representations of the biopsy datasets are visualized in two dimensions using PCA. We observed that the feature space learned by the CLAM model from resections remains visibly separable among the distinct subtypes when it is adapted to biopsy slides for both NSCLC (f) and RCC (g). A high-resolution version of these biopsy whole slides and heatmaps may be viewed our interactive demo (http://clam.mahmoodlab.org).
