Abstract
Since the discovery of the yellow fever virus in 1901, thus far, two hundred nineteen viral species are recognized as human pathogens. Each year, the number of viruses causing infections in humans increases, triggering epidemics and pandemics, such as the current COVID-19 pandemic. Pointing to bats as the natural host, in 2019, a genome highly identical to a bat coronavirus (COVID-19) spread all over the world, and the World Health Organization (WHO) officially confirmed it as a pandemic. The virus mainly spreads through the respiratory tract, uses angiotensin-converting enzyme 2 (ACE2) as a receptor, and is characterized by symptoms of fever, cough, and fatigue. Antivirals and vaccines have provided improvements in some cases, but the discovery of a new and diverse variety of viruses with outbreaks has posed a challenge in timely treatments for medical scientists. Currently, few specific antiviral strategies are being used, and many of the effective antiviral drugs and reported active molecules are under vital exploration. In this review, with the details of viral diseases, we summarize the current attempts in drug development, epidemiology, and the latest treatments and scientific advancements to combat the COVID-19 epidemic. Moreover, we discuss ways to reduce epidemics and pandemics in the near future.
Keywords: Viruses, Covid-19, HIV, SARS, MERS-CoV, 3D structure analysis
1. Introduction
Parasites as viruses are not only responsible for a high rate of human morbidity and mortality but are also responsible for epidemic and pandemics (Choudhary and Dar, 2018). International Committee on Virus Taxonomy (King et al., 2012) have classified viruses according to the genome and morphology. The genetic material can be single or double-stranded DNA or RNA, for single it can be plus or minus sense. Retroviruses are in a different group including DNA and RNA-encoded viruses. Henceforth, they are classified into seven classes (Caprari et al., 2015). Viruses are responsible for skin diseases and are responsible for primary, secondary, or systematic infections which may lead to sometimes formation of tumors and sometimes only inflammation (Sterling, 2016). The most frequent viruses are airborne common cold viruses that are not life-threatening but highly contagious, while influenza viruses may result in fatalities (www.CDC.gov/measles/) (Corstjens et al., 2016). However, viruses as Adenovirus, Coronavirus, Hantavirus are not common but have a great impact on public health (Stollenwerk et al., 2008).
Viruses are responsible for different diseases and result in the outbreak of epidemics and sometimes pandemics as COVID-19 is the present example (Table 1 ). Some of the diseases caused by different viruses are discussed below.
Table 1.
Some known Pandemic and Epidemic.
| Pandemic/Epidemic | Origin | Year |
|---|---|---|
| Philadelphia yellow fever epidemic | Philadelphia, the United States | 1793 |
| Flu pandemic | St. Petersburg Russia | 1889–1890 |
| American polio epidemic | New York City | 1916 |
| Spanish Flu | China | 1918–1920 |
| Asian Flu | Singapore | 1957–1958 |
| AIDS pandemic and epidemic | West Africa | 1981 |
| H1N1 Swine Flu pandemic | Mexico | 2009–2010 |
| Middle East respiratory syndrome coronavirus (MERS-CoV) | Middle East | 2012 |
| West African Ebola epidemic | Sudan | 2014–2016 |
| Zika Virus epidemic | South and Central America | 2015 |
| Coronavirus Covid-19 | Wuhan China | 2019 |
The infectious diseases that are transferred from animals (vertebrate) to humans and from humans to animals are called Zoonotic diseases. All kinds of microbes are responsible for this type of disease. The impact of these diseases has increased considerably in the last decades although vaccination and effective therapies have been implemented for their prevention. But still, there is a need for effective investigation and control in the human-animal–ecosystem interface (Wang and Crameri, 2014). Gastroenteritis may be due to different pathogens like bacteria, parasites, and viruses. It is now known that human caliciviruses are mostly the main reason for diarrhea for people around the world (Patel et al., 2009).
Viruses as Noro, rabies, human papilloma, Epstein-Barr, herpes simplex, and HIV are transmitted by the oral cavity. Henceforth, infections are manifested by using oral samples (Corstjens et al., 2016). However, virus transmission is not caused by only one route, but different viruses take different routes to enter the host which may be as simple as only by sneezing or coughing. Noroviruses are spread by the fecal-oral route and its spread may also be attributed to both environmental surfaces and aerosolization with outbreaks in closed settings (Patel et al., 2009).
Arenaviridae and Filoviridae are the classes of viruses causing hemorrhagic fevers. However, hemorrhagic symptoms may be associated with some Flaviviruses and some Bunyaviridae (Yuill, 2018). Different taxonomical groups of viruses are responsible for viral hemorrhagic fever (VHF), but they all produce systematic and fatal diseases such as the Ebola virus, other viruses are Marburg, Lassa, the South American arenaviruses, yellow fever, Crimean-Congo anaq Rift Valley fever viruses. All of these viruses attack the host immune system and change the cells responsible for antiviral response (Geisbert and Peter, 2004).
Acute respiratory viruses are also one of the major reasons for morbidity and mortality in humans and worldwide. Many of these viruses attack young infants, the elderly, and immune-compromised individuals (Osterhaus, 2008). It is caused by avian paramyxovirus serotype-1 viruses and affects poultry and is a threat to existing or developing poultry industries. This disease may infect humans but human to human spread has not been reported so far (Alexander, 2000). Tick-borne diseases of humans such as encephalitis and Crimean-Congo hemorrhagic fever in recent years have tremendously increased and may pose a problem to public health in the future through global warming and suggested to be responsible for such rise in this kind of diseases, however, other environmental factors cannot be ignored in such diseases (Estrada-Peña and De La Fuente, 2014). The current review focuses on the viruses, which have emerged from animal sources and have played a role in emerging viral diseases. It also focuses on the current approach to control the epidemics leading to pandemics and current therapies for the recent pandemic. A few of the viruses are discussed below.
2. Types of viruses
2.1. Paramyxoviruses
Paramyxoviruses are a large group of viruses and include Hendra, Nipah, parainfluenza viruses types 1–5, Newcastle disease, mumps viruses, Yucaipa (Dinter et al., 1964), and Nariva (Walder, 1971) viruses. Measles, canine distemper, and rinderpest viruses are found to be structurally like the paramyxoviruses and hence can be accepted in this group. Paramyxoviridae family, whose member Genus's includes Rubulavirus, Avulavirus, Respirovirus, Pneumovirus, and Metapneumovirus. Bat viruses as human parasites have also been perceived in huge quantities, including bat lyssaviruses, parainfluenza viruses, hantaviruses, hepaciviruses, and pegiviruses (Wang and Crameri, 2014). Besides, a large number of other viruses as paramyxo, corona, astro, adeno, and herpes have also been reported (Wang and Crameri, 2014).
The nucleocapsid is surrounded by lipid envelop and envelop is formed by attaching to host cell plasma (Lamb and Parks, 2012). The family is split into two subfamilies, Paramyxovirinae and Pneumovirinae (Fauquet et al., 2005), with multiple genera. The genera of Paramyxovirinae include Rubulavirus (several species of human parainfluenza viruses and the mumps viruses), Avulavirus, Morbillivirus, Respirovirus, Henipavirus, and the later containing Pneumovirus genera and Metapneumovirus. Serious respiratory syncytial viruses (RSV) diseases are caused by Pneumovirinae and are the leading cause of pneumonia in children across the world. The classification of respiratory syncytial viruses is problematic as the polymerase gene is similar to Filoviruses than to the Paramyxovirinae. The classification of these viruses is challenging as the gene (polymerase) is associated more with those of the Filoviruses. Human RSV is the most important cause of pneumonia in infants and children worldwide (STRAUSS and STRAUSS et al., 2008). New viruses are being discovered in wild animals continuously and constantly the family of Paramyxoviridae viruses are increasing with a lot of uncharacterized viruses (Maclachlan and Edward, 2010).
Coronaviruses including (severe acute respiratory syndrome virus, Middle East respiratory syndrome virus), Filoviruses (Ebola and Marburg viruses) (Choppin and Compans, 1975; Wang and Crameri, 2014), are RNA viruses that are single-stranded and often create infections of the respiratory tract. RNA viruses quickly adapt to changes due to their high rate of errors by viral enzymes responsible for its genomic replication. These viruses are responsible for many emerging or re-emerging diseases while different factors as genetic differences in viruses and certain environmental factors also play a role in the emergence of diseases (Nichol et al., 2000). Viruses such as human metapneumovirus and 2 different human coronaviruses were with humans for centuries, but interspecies transmissions have led to the discovery of different new viruses from other mammalian or avian reservoirs, such as avian influenza viruses, severe acute respiratory syndrome (SARS), Corona and Nipah virus. This has resulted in severe diseases and outbreaks such as epidemics and pandemics also (Osterhaus, 2008).
2.2. Menangle virus and reovirus
Menangle virus belongs to the Rubulavirus genus (Paul, 2010) and has a spikes layer on the surface of the virus. These viruses are related to the Tioman virus and Melaka (Chua et al., 2007; Kohl et al., 2012). Nipah virus, Tioman virus, and reovirus (Pulau virus) were isolated from fruit bats (Pritchard et al., 2006). These reoviruses (respiratory enteric orphan viruses) nonenveloped viruses with double-stranded RNA taxonomically belongs to the group Reoviridae (Chua et al., 2007).
2.3. Pegiviruses
Hepaciviruses and pegiviruses are from the family Flaviviridae and belongs to closely related genera (Bunney et al., 2017). Recently, the hepaci-/pegi-like virus has been described in sharks and henceforth its existence in non-mammalian species indicates that further genera remain to be defined in the future (Smith et al., 2016).
2.4. Lyssaviruses
The genus Lyssavirus belongs to the family Rhabdoviridae and includes viruses causing the fatal encephalitis rabies (Zhu et al., 2016). These viruses are bat-borne and play an important role in the epidemiology of rabies and some rabies-like viruses. Transmission of viruses by a bat is mostly by bite although other routes may also be possible (McCOLL et al., 2000).
2.5. Herpesvirus
More than 200 herpesvirus species have so far been identified and in humans the dsDNA viruses which are wrapped in icosahedral capsid cause diseases as cold sores, chickenpox, and genital herpes. These species also have viruses that may cause cancer (Riaz, 2017). It now includes 90 species (Davison, 2010). These viruses suppress the host immune response (Ryu, 2017).
2.6. The Adenoviridae
The family Adenoviridae currently comprises five genera in which Mastadenovirus, comprising viruses infecting mammals only. All adenoviruses have a similar morphology however the genomic organization is different between viruses of different genera (King et al., 2012).
2.7. Human astrovirus (HAstV)
This virus is responsible for causing diarrhea and may lead to some times hospitalization due to it (Jeong et al., 2012). Initially, the Astroviridae family consisted of two genera based on the host's origin: Mamastrovirus (MAstV) and Avastrovirus (positive-sense single-stranded RNA), which are found to cause infection in mammals and avians. HAstV infection causes diarrhea with vomiting, fever, and abdominal pain. Vomiting is more pronounced in the case of rotavirus or calicivirus infection (Bosch et al., 2014).
2.8. Hantaviruses
Hantavirus belongs to a family Bunyaviridae responsible for hemorrhagic fever which was firstly reported in 1951–1953 among UN troops. In 1981 it was established that these viruses have a specific rodents host and haven't arthropod vector which other viruses in the family possess. Henceforth, a new genus “hantavirus” was introduced in the Bunyaviridae family including viruses causing hemorrhagic fever with renal syndrome (HFRS). Now it is well established that in 1993, a serious respiratory problem along with cardiopulmonary syndrome led to the discovery of Hantavirus disease. More than 21 species are now included in the Hantavirus genus (Mir, 2010). The most lethal HFRS-associated Hantavirus is the Hantaan virus in Asia whereas in Europe it is the Puumala virus and Dobrava virus. Andes virus (ANDV) and Sin Nombre virus (SNV) are the most lethal of the type (Hammerbeck et al., 2009).
2.9. Human parainfluenza viruses (HPIV)
Discovered in the 1950s belong to the Paramyxoviridae family, genetically and antigenically divided into 1 to 5 serotypes with two genera Respirovirus and Rubulavirus (Parija and Thomas, 2019). Respirovirus are responsible for lower respiratory infections and are closely related to megamyxoviruses and metapneumovirus (Henrickson, 2003) and other closely related viruses that cause influenza-like symptoms or pneumonia (Brenda, 2020).
H5N1 strains of avian influenza A virus was responsible for a large number of deaths since its outbreak in Southeast Asia in 1997 (Proença-Módena et al., 2007). It is believed that virus-encoded nonstructural protein (NS1) is synthesized in infected cells and results in reduction of the host interferon-β mRNA (Krug, 2006).
2.10. Arboviruses
The origin of viruses is very difficult to assess, however, pieces of evidence have suggested that yellow fever, chikungunya fever, and Zika fever are African based and it is believed that dengue fever is also African in origin (Powell, 2018). These are of blood-feeding arthropods-borne viruses, chiefly insects (flies and mosquitoes), and arachnids (ticks) which affect a larger number of people by spreading widely. Air travel and mosquitos as vectors contribute significantly to its spread. Dengue, yellow fever, chikungunya, and Zika viruses are examples of such viruses (Weaver et al., 2018). Arboviruses have been affecting humans for centuries and cause considerable complications inpatients out of these four are found to be widely and rapidly spread as the viruses causing yellow fever, dengue fever, chikungunya fever, and Zika fever (Powell, 2018). Arbovirus classification is not included in the current classification of the viral system. Bunyaviridae, Togaviridae, Flaviviridae, and Reoviridae are the families of viruses in the current classification system that has arbovirus members included in it.
Arenaviridae and Filoviridae are families causing hemorrhagic fevers however, some of the flaviviruses and Bunyaviridae may also be linked with symptoms of hemorrhagic (Yuill, 2018). A neurotropic flavivirus as the West Nile virus belongs to the family of Flaviviridae (Suthar et al., 2013).
2.11. Anello viruses
Human anelloviruses includes Torque Teno virus (TTV), and TTV-like mini virus (TLMV), are found to be commonly present in chronic hepatitis, cirrhosis, and hepatocellular carcinoma patients, for a long period.but have not been found to be responsible for liver diseases. However, no diseases so far with TTV has been reported in humans (Naik et al., 2020). In most of the human plasma samples small DNA single-stranded mini-viruses as TTV and TTV-like mini-virus (TLMV) were found to be infecting man (Thom et al., 2003).
The only known single-stranded circular DNA found in humans so far is the TTV. Also, another virus that resembles the TTV is the TLMV. TLMV does not have to envelop but its density is similar to TTV (Takahashi et al., 2000).
2.12. Coronaviruses
Coronaviruses belong to the family Coronaviridae, which is further composed of two subfamilies, the coronaviruses, and the toroviruses. Toroviruses are responsible for causing enteric diseases in cattle and possibly in humans. Coronaviruses serologically are further divided into three groups (I to III). Group I coronaviruses include not only animal pathogens but also human coronaviruses HCoV-229E and HKU1 (Weiss and Navas-Martin, 2005). Coronavirinae is further divided into four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (Cui et al., 2019). This classification is based on genomic structures and phylogenetic relationships. The transmission of alphacoronaviruses and beta coronaviruses is found to be limited to mammals, while the other classes also infect birds. These viruses in humans cause respiratory diseases such as, SARS coronavirus and Middle East respiratory syndrome coronavirus (Forni et al., 2017; Kumar et al., 2020). The coronavirus virion contains spikes, which are multifunctional molecular machines and assist in the entrance of coronavirus into host cells. Spike has two subunits, S1 and S2; S1 binds to the receptor on the surface of the host cell, while S2 transfers viral and host membranes into the host (Li, 2016). SARS-CoV and MERS-CoV have shown different behavior compared to SARS-CoV-2 (Guarner, 2020).
By the end of 2002, severe acute respiratory syndrome (SARS) began to spread rapidly around the world, while its outbreak originated in China (Huang, 2004). The SARS coronavirus is responsible for severe acute respiratory syndrome (SARS) disease.
A betacoronavirus is also responsible for Middle East respiratory syndrome coronavirus (MERS-CoV), a zoonotic viral respiratory illness first reported in Saudi Arabia in 2012 (Al Awaidy and Khamis, 2019). One of the symptoms of MERS is also pneumonia, and the virus is transmitted through contact with infected camels or people. In MERS-CoV-endemic countries, social distancing isolation and management of the patients are necessary to prevent outbreaks (RESEARCH HIGHLIGHT KAIMRC, 2019).
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the positive single-stranded RNA virus that is contagious in humans and is the reason for the ongoing pandemic of COVID-19 disease. The new strain of coronavirus was discovered in December 2019 in Wuhan, China. This virus is also included in the same family that caused SARS and MERS (Parry and Peterson, 2020; Yanping, 2020). It has been reported that this virus is sensitive to high temperature (heat) and can be inactivated by a few lipid solvents (Cascella et al., 2020). SARS-CoV-2 has been included in the beta coronavirus group and is found to resemble bat coronavirus HKU9-1. Its spike protein is found to interact strongly with the human ACE2 receptor (Tahir et al., 2020), with its pathology resembling those of SARS and MERS coronavirus infections. Moreover, it has been reported that an injury in the liver could be caused by either SARS-CoV-2 infection or drugs. A liver biopsy specimen of a COVID-19 patient was found to have mild microvascular steatosis and portal activity. Pneumonia was found to progress rapidly with some differences between the left and right lungs. It was suggested that the use of corticosteroids in severe patients could prevent acute respiratory distress syndrome development (Xu et al., 2020). This virus was found to be transmitted from person to person in the hospital and among family members and from infected travelers to other regions (Chan et al., 2020).
It was also suggested that no intrauterine infection was found in women with COVID-19 pneumonia in late pregnancy (Chen et al., 2020). RNA viruses adapt very quickly to environmental changes, and resistance to drugs is also very prevalent. This adaptability and resistance to drugs are due to mutations in the viral genomes and, hence, the main reason for emerging diseases (Holmes and Rambaut, 2004). It was observed from the chest CT of recovered COVID-19 patients whose lung abnormalities were maximal after 10 days of initial onset of symptoms (Pan et al., 2020).
People with diabetes, hypertension, or other chronic diseases are severely affected by SARS-CoV-2. It has been reported that coronavirus gains entry to infect the cells by using Angiotensin Converting Enzymes 2 (ACE2) and since diabetic patients utilize ACE inhibitors, it further increases the expression of ACE2 thereby facilitating the viral uptake and increasing the severity of infection in them. However, it is further suggested that using the renin-angiotensin system blockers may be helpful in decreasing the disease risk but it needs further investigation (Ma and Holt, 2020).
3. Computational outcomes and challenges
With the conjunction of bioinformatics and computational methods with publicly available phenotypic data of host responses to drugs and pathogens, it is now possible to identify new lead molecules as drug candidates for cancers and to identify numerous therapeutic agents for different parasitic diseases (Law et al., 2013). Computational tools have found applications for not only noninfectious diseases but also infectious diseases. However, fewer studies are performed on parasitic diseases (Marhöfer et al., 2011; Shamshad et al., 2020).
Among different target the two coronaviral proteases, as papain-like protease (PLpro) and 3C-like protease (3CLpro) have gained importance for targeting antiviral drugs as they play a role in the replication of the virus (Báez-Santos et al., 2015). Targeting PLpro is more challenging as it may also inhibit host-cell deubiquitinases (Ullrich and Nitsche, 2020) while 3Clpro (also referred to as Mpro) is one of the best-characterized targets. 3Clpro cleaves polyproteins on 11 sites and cleaves its N- and C- terminal sites by P4–P1 and P1’ sites recognition (Muramatsu et al., 2016). Inhibitors against 3Clpro may be selective as no human protease with the same cleavage is present (Zhang et al., 2020).
We selected 3Clpro protease proteins from protein data bank [https://www.rcsb.org/] representing 7C8T.pdb for SARS-CoV-2, 4RSP.pdb for MERS-CoV, and 2GX4.pdb for SARS-CoV with Escherichia coli as an expression system. 3Clpro proteinase is a member of the cysteine protease family, having chymotrypsin fold which is similar to picornaviruses (Huang et al., 2004). It is a homodimer having one active site per in each unit (Muramatsu et al., 2016).
It consists of three domains having active site in the domain I and II which have antiparallel β-barrel structures with the unconventional catalytic dyad Cys145 and His41 buried inside the active site. Several crystal structures of 3CLpro apo and with inhibitors have revealed common features with two domains from residues 1–184 and one helical domain with residues 201–303 (Fig. 1 ). However, the additional 100 residues in the III domain was found to be absent from the picornavirus protease (Liang, 2006). Moreover, domain III with five α-helices plays a role in the dimerization of the enzyme and is connected to domain II via a long loop. The 3Cl protease which is not present in the picornavirus is important for the dimerization and enzymatic activity. The N-finger having N-terminal amino acids is also important as its deletion leads to failure in enzymatic activity. N finger region is clasped between domains II and III of the one monomer unit and domain II of the other monomer as shown in Fig. 1 (Kneller et al., 2020). Active residues are divided into S1–S6 subsites with catalytic dyad His41–Cys145 present in the S1 subsite. S2 and S4 (Fig. 1) have hydrophobic residues, while S5 and S6 are away from the catalytic dyad.S5 and S6 may contribute less to the substrate-binding (He et al., 2020). S1 is the substrate-binding site in one subunit confers specificity for the P1-Gln substrate residue on the enzyme. The conservation has been reported for the amino acid sequence Leu-Gln-Ser or Leu-Gly-Ala (P2–P1–P1’) (Pillaiyar et al., 2016). The P4, P3, P2, and P1 residues interact with the substrate-binding sites (Muramatsu et al., 2016).
Fig. 1.
a) Aligned 3D structures of the selected proteins brown is 4rsp.pdb, blue 2gx4.pdb and green 7c8t.pdb, b) 3D structure in which red color indicates helics, green β turns, purple ɤ turns brown β hair pin region, c & d) structure according to the showing the substrate binding region.
It has been suggested that in P1 presence of glutamine is important while for the P2, P3, and P4 positions, leucine, basic residues, small hydrophobic residues are preferred. In the same way, small residues are essential for P1’ and P2’ positions (Jo et al., 2020).
It has been reported that on ligand binding shift to about 2.4 Å is observed for P2 residues 45–50 and the β-hairpin loop residues (P3–P4) 166–170, loop residues 190–194 (P5) was found to move closer to the moves closer to the β-hairpin loop residues. Since the residues of P2, P5, and C terminal tail were reported to be flexible hence can accommodate various chemical entities at the P2–P5 sites (Kneller et al., 2020).
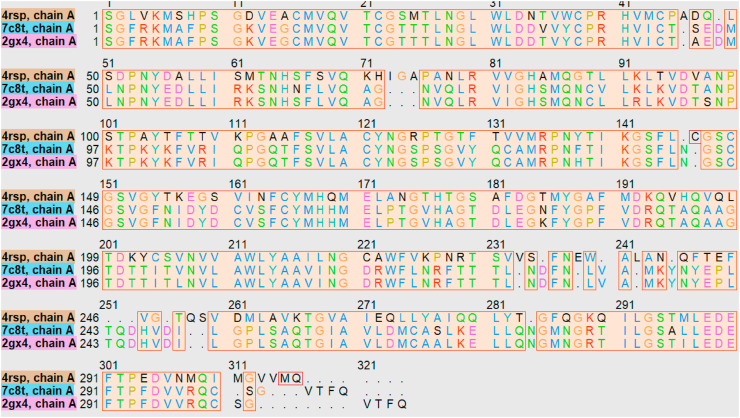
Henceforth in this context, the protease proteins 7C8T.pdb, 4RSP.pdb, and 2GX4.pdb were downloaded from the protein data bank [https://www.rcsb.org/] and were used to analyze the 3D similarity of the selected structures using Chimera (Pettersen et al., 2004). 7C8T.pdb was selected as reference protein and the three proteins were overlapped as shown in Fig. 1 using matchmaker in Chimera with Needleman-Wunsch (BLOSUM-62) and the RMSDs were calculated (Table 2 ). The alignment was created by the superposition of the protein structures as shown in Fig. 2 using chimera and it showed the superposition across all 292 fully populated columns having structural similarity score (cutoff 5.0): 22.303 and Q score 0.767. The sequence length was 306, 304, 306, with RMSD values for the fully populated columns in alignment as shown in Table 3 . So, in the future, there is a huge possibility that single antiviral drugs can inhibit the three viruses, simultaneously. May be an ensemble of three proteins can be used for docking of the compounds and can be used in the quest for antivirals that can simultaneously inhibit the three proteins (Scheme 1 ).
Table 2.
3D similarity using 7C8T.pdb as a reference protein.
| Chain A | 7C8T with 4RSP | 7C8T with 2GX4 |
|---|---|---|
| Sequence alignment score | 953.1 | 1541.8 |
| Pruned atom pairs | 0.954 (252 atoms) | 0.546 (297 atoms) |
Fig. 2.
Sequence alignment for the proteins 4RSP.pdb, 2GX4.pdb and 7C8T.pdb. Colored boxes indicate the similarity with the 3D structures. Purple shows highly conserved residues, while completely conserved are shown in red.
Table 3.
Superposition across all 291 fully populated residues in the alignment represented by RMSD.
| 7C8T with 4RSP | 7C8T with 2GX4 | 4RSP with 2GX4 | Overall RMSD |
|---|---|---|---|
| 1.344 | 0.655 | 1.245 | 1.123 |
Scheme 1.
Schematic view of the transmission of viruses and illustration of 3D similarity of viral enzymes
4. In silico drug design
It has been reported that most antivirals that had been approved until 2006 were based on natural products. Although computational tools have been successful in guiding and accelerating the drug development process, they are still too immature to tackle the current viral threats on time (Mirza, 2019). Several viral diseases have been extensively studied computationally but, but there has yet to be a breakthrough in the quest of antivirals, as in the case of HCV (Kirchmair et al., 2012).
In silico docking studies were carried out that showed 18 flavonoids with the ability to strongly bind with influenza haemagglutinin stems of various subtypes, which are the target for antibodies (Mathew et al., 2018). Target-based virtual ligand screening was performed using 21 targets against compound libraries including the ZINC drug database and natural products and 78 commonly used anti-viral drugs. Through these studies, potential inhibitors for different targets were predicted (Wu et al., 2020). SARS-CoV-2 protease in complex with an inhibitor was used to screen approved drugs and drugs in clinical trials (Wang, 2020). Through docking studies, candidate drugs were identified, including carfilzomib, eravacycline, valrubicin, lopinavir, and elbasvir. Carfilzomib was a promising inhibitor against SARS-CoV-2 (Wang, 2020). The bioactive compounds from the medicinal plant could dock to SARS-CoV-2 protein, and compounds such as kaempferol, quercetin, demethoxycurcumin, apigenin-7-glucoside, naringenin, oleuropein, curcumin, catechin, and epicatechin-gallate were found to be effective inhibitors against SARS-CoV-2 (Khaerunnisa et al., 2020).
Docking studies also showed that Nigellidine and α-Hederin can be potential inhibitors of the SARS-CoV-2 virus and medicinal use of sativa against coronavirus infection requires further research and attention (Salim and Noureddine, 2020). Molecular docking studies on 7 proteins of SARS-CoV-2 were performed, and a compound present in green tea, epigallocatechin gallate was found to be an effective binder of protein (Khan, 2020). Molecular docking studies on 80 flavonoid compounds were performed, and hesperidin, rutin, diosmin, apiin, diacetylcurcumin, and others were found to be inhibitors of SARS-CoV-2 (Adem et al., 2020). In another study coumermycin, trovafloxacin, aclarubicin, raltegravir, etoposide, epirubicin, nystatin, talniflumate, idarubicin, itraconazole, maraviroc, and nelfinavir were found to be inhibitors of SARS-CoV-2 (ARULANANDAM, 2020).
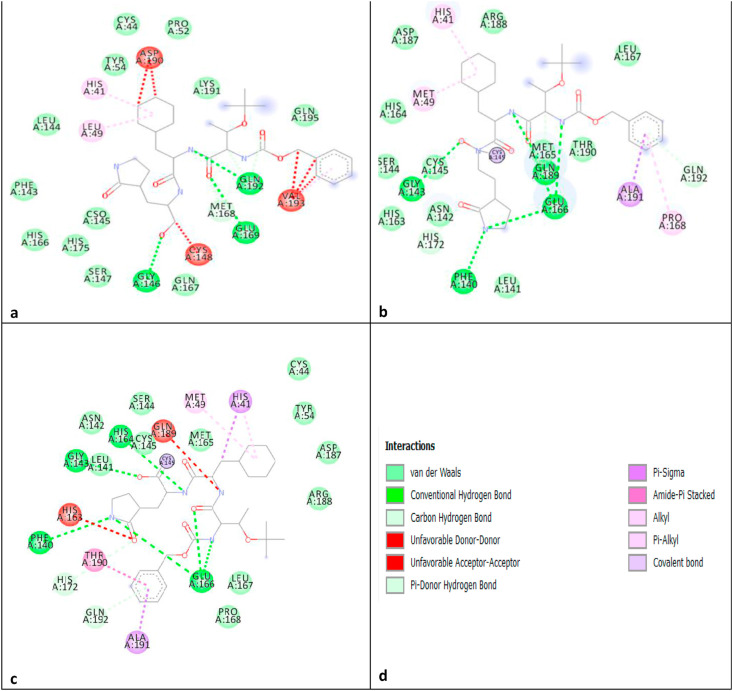
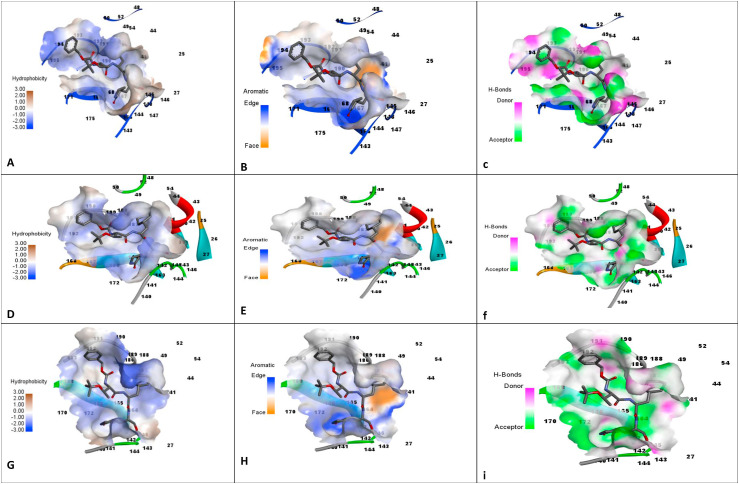
The overlapped three proteins were used for the study of the interaction pattern of proteins with the deposited inhibitor TG-0205221. Moreover, MERS protease 4RSP.pdb was also used to study the interaction pattern of the same inhibitor in its binding cavity using DS. Fig. 3 shows the interaction profile of the three proteins which indicates that conserved residues involved in the three proteins making important interactions with the similar ligand. Henceforth, it is highly possible that a single ligand may be potent against the three targets. Fig. 4 shows the binding site characteristics (hydrophobicity, aromaticity, and hydrogen bonding) of the three proteins. It can be used to design in future the ligand for the three proteases.
Fig. 3.
a) 4RSP.pdb, b) 2GX4.pdb, c) 7C8T.pdb, d) with blue, black or green dashed line with arrow at H bond acceptor.
Fig. 4.
Binding site characteristics of hyphobicity, aromaticity and H bonding for the proteins, a–c: 4RSP.pdb, d–f: 2GX4.pdb, g–i:7C8T.pdb.
5. Treatments
Virology has helped in the understanding of the immunological processes and has shaped the current immunological systems. Animal and human models of viral infections played a pivotal role in the discoveries of immunological processes (Abdel-Hakeem, 2019). Immunoprophylaxis is of two types, active and passive. Active prophylaxis involves stimulation of the body's immune system by administering viral preparation in the form of attenuated life, killed or recombinantly produced antigens. In passive prophylaxis antibody from another host is administered to protect patients. Many viral diseases are controlled by good sanitation practices. Using antiviral therapies which are of three types one which inactivates the virus the second inhibit its replication and the third one boosts the host immune response (Goldenthal et al., 1996).
To reduce epidemics world-wise policies have been successful to some extent and several infectious diseases have been eradicated or controlled due to vaccination programs. In most cases, eradication was possible because the viral structure comprised of a single strain as Smallpox. Vaccination in the case of Measles also proved to be effective for its eradication (Corstjens et al., 2016).
Chikungunya virus an alphavirus has 3 genotypes: Asian, West African, and Eastern/Central/South African. However, no vaccine for Chikungunya is available, and antibody reaction on one genotype is effective against another is still not clear (Goo et al., 2016).
6. Available therapies
Most viral diseases self-resolve and do not require any specific treatments. However, antiviral drugs for three groups of viruses, herpes, hepatitis, and influenza are in clinical trials currently apart from HIV drugs (Razonable et al., 2020). Idoxuridine was the first antiviral agent approved in 1963; since then, 90 antiviral drugs having 13 different functions have been approved for the treatment of viral infections, including HIV, hepatitis B and C, influenza, herpesvirus, varicella-zoster, human cytomegalovirus, respiratory syncytial virus, and human papillomavirus (Clercq and Li, 2016). The majority of antiviral drugs approved during the 1987–2017 period, were for HIV-1, and the second most abundant antiviral drugs were for HCV. Four were approved for HPV infection and influenza and five for respiratory viruses, while one was approved for a respiratory syncytial virus (RSV) (Chaudhuri et al., 2018; Clercq and Li, 2016). It was reported that a combination of statin/caffeine as preventative therapy was as effective as oseltamivir and ribavirin. Hence this could be used as an antiviral formulation in the future. It has also been suggested that statins could be helpful as antiviral therapy and can have significant effects on influenza outcomes (Bkhaitan, 2017; Liu et al., 2009).
7. Drug development for the current pandemic
The rapid spread and emergence of COVID-19 have urged the requirement for an effective therapeutic agent against it. The National Health Commission (NHC) has so far recommended IFN-α, lopinavir/ritonavir, ribavirin, chloroquine phosphate, and arbidol as anti-viral therapies. Many approaches have been used to target SARS-CoV-2 progression such as inhibition of its fusion/entry, replication, inflammation, plasma treatment, vaccines as well as traditional and FDA approved medicines. Numbers of clinical trials are also in progress to determine the safety and effectiveness of candidate drugs (Haiou et al., 2020).
Various categories of drugs are being investigated for this pandemic, including favipiravir or avigan, which was found to be effective in China against COVID-19 infections. Tocilizumab will be on trial by Roche to determine its outcome in patients with COVID-19 (Live Science Staff, 2020). Initially, a compound named 3a was found to effectively treat several viruses in lab experiments, even a type of coronavirus (Cho et al., 2012). Now, the derivative of that molecule, remdesivir has been used to treat patients with coronavirus infection in the hope it can reduce the intensity of COVID-19 (Joseph, 2020). Remdesivir is a nucleotide analog that is intravenously given to patients, and no adverse effects were observed after the infusion. However, randomized controlled trials are required to assess the safety and efficacy of the drug for treating 2019-nCoV infection (Lofy et al., 2020). Remdesivir and IFN-β were found to have better antiviral activity compared to LPV and RTV in vitro. Moreover, in vivo evidence has also demonstrated that remdesivir has the potential to treat MERS-CoV infections (Sheahan et al., 2012). It has been reported in-vitro that the drug combination of remdesivir and chloroquine was effective against SARS-CoV-2. Moreover, HIV protease inhibitors lopinavir and ritonavir were also found to be effective against COVID-19 patients in Korea. A combination of Chinese and western medicines was also found to be effective (Guo et al., 2020). In a current report, 12 FDA-approved drugs were suggested to serve as therapeutics for MERS-CoV or COVID-19 therapy. The cardiotonic drugs as monotherapy or combined with remdesivir or with other therapeutics were considered best for therapy (Ko et al., 2020).
The in vitro activities of the antimalarials hydroxychloroquine and chloroquine have been reported against COVID-19 (Table 4 ). However, a small study in China showed no progress in the recovery of patients with COVID-19. Moreover, the side effects and toxicity of anti-malarial drugs should be addressed in critically ill patients (Yazdany and Kim, 2020). Monoclonal antibodies have been designed for SARS-CoV and not only are used in clinical practice but also have been applied in SARS patient therapy. These studies may be helpful in vaccine development against SARS-CoV-2 (Tsunetsugu-yokota et al., 2006). MicroRNAs serve as regulatory RNAs and may play a role in the pathology of several respiratory viruses; therefore, they may serve as therapeutic agents in respiratory viral infections (Tahamtan et al., 2016).
Table 4.
Drugs under investigation for the current Pandemic.
| Structure | Name | Type of drug |
|---|---|---|
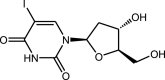 |
Idoxuridine | Nucleoside derivative |
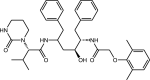 |
Lopinavir | Dicarboxylic acid diamide |
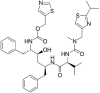 |
Ritonavir | L-valine derivative |
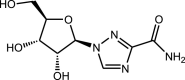 |
Ribavirin | Nucleoside analog of ribofuranose |
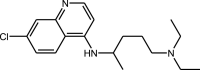 |
Chloroquine | 4-aminoquinolines |
 |
Arbidol | Indole core |
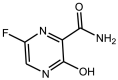 |
Favipiravir or Avigan | Pyrazinecarboxamide derivative |
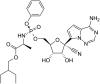 |
Remedesivir | Ribonucleotide analog |
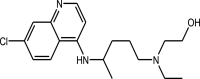 |
Hydroxychloroquine | 4-aminoquinolines |
8. Vaccine and drugs in clinical trials
Successful clinical trials aide in the approval of new drug product as the trials confirms the safety and efficacy of the new product. However, the lapses in the procedure could be improved and modernized. Current COVID-19 has paved the way for the improvement in the current methodology of the clinical trials. The improvement in the system may in the future help practitioners patients for the rapid and on-time launch of the drug or vaccine (van Dorn, 2020).
For the current pandemic, two types of approaches are used to prevent the disease one involves the use of antivirals and the second one involves immune modulators. In one of them using only randomized controlled trials for more than 100 participants use different antivirals as remdesivir, lopinavir, chloroquine/hydroxychloroquine, favipiravir, etc have been reported. Moreover, immune modulators as dexamethasone, convalescent plasma, azithromycin, interferons, anakinra, leronlimab, etc have also been reported (Robinson, 2020).
Initially, interferons nebulization and antiviral drugs were in practice to circumvent the disease. However, remdesivir has been so far found to be effective against the disease. Different drugs also went through a number of trials such as hydroxychloroquine, azithromycin, the antiviral compounds ritonavir and lopinavir, and tocilizumab, chloroquine, ivermectin, favipiravir, umifenovir, and oseltamivir. However, only 5 and 4 trials for the serine protease inhibitors were reported (Idda et al., 2020).
Vaccine development is not an easy task and is not only expensive but also very complex and risky. It is risky as many vaccines may fail in the initial or final stages (Douglas and Samant, 2013). More than eighteen and nine years have been passed after the first case of SARS and MERS was observed. No vaccine has been so far approved for the prevention of corona based diseases. There are varied reasons for the lack of a vaccine against these diseases. The most important reason may be the lack of interest in developing vaccines for a disease that has was found to be a geographically centralized and low number of cases. However, it has been reported that most of the vaccine were in clinical phase 1 for the SARS and MERS (Padron-Regalado, 2020).
In the current scenario of the corona pandemic, more than 150 vaccines are in developmental stages to circumvent the pandemic in record time. For this purpose, a virus in an attenuated, killed, or weakened state is being utilized while other parts of the virus as protein/fragments are also in the vaccine developmental stages. Some of the tested vaccines against corona are adenovector vaccines in which a fragment of DNA is inserted in weakened adenovirus and is undergoing three ensemble trials. In another study, SARS-COV-2 spiked protein is used as a vaccine with weakened adenovirus as a vector and is in phase 2 and 3 stages.
In the same way, mRNA fragments of the virus will be injected into the human cells for creating the viral protein which may train the immune system for recognizing the virus. This will be the first of the technology if approved and is currently in the third phase. Trojan horse is used in one of the studies as a vector for the coronavirus vaccine. CoronaVac which is an inactivated form of coronavirus is also in phase 3. Sinopharm trial vaccines are also using an inactivated form of coronavirus and are the first vaccine given to civilians. Bacillus Calmette-Guerin (BCG) vaccine may also boost immune response in corona disease (MCKEEVER, 2020). In older adults with the mRNA-1273 vaccine, adverse effects were reported to be mild and are in phase 3 of the trial (Anderson et al., 2020). Table 5 lists some of the vaccines and drugs in clinical studies so far for the COVID-19 pandemic.
Table 5.
Drugs that have completed clinical trials and vaccines in clinical trials.
| T.P* | Drugs and combinations | T.P | Vaccine |
|---|---|---|---|
| 4 | Danoprevir + Ritonavir | 4 | Bacille Calmette-Guérin (BCG) Vaccine |
| 4 | Ganovo + Ritonavir | 4 | Polio vaccine |
| 3 | Favipiravir | 3 | Measles-Mumps-Rubella Vaccine |
| 3 | Hydroxychloroquine | 3 | Candidate Coronavirus Disease Vaccine (ChAdOx1 nCoV-19) |
| 3 | Baricitinib | 3 | IMM-101 immune stimulating therapy for cancer patients |
| 3 | Hydroxychloroquine + Azithromycin | 3 | Reconstituted vaccine containing VPM1002 (Mycobacterium bovis) |
| 3 | Remdesivir | 3 | Convalescent immune plasma treatment |
| 2 | Chloroquine diphosphate | 2 | Recombinant Novel Coronavirus Vaccine (Adenovirus Vector) |
| 2 | Tirofiban injection associated with acetylsalicylic acid IV + Clopidogrel + fondaparinux | 2 | Inactivated SARS-CoV-2 vaccine |
| 2 | Lopinavir/ritonavir + Ribavirin + interferon beta-1b | 2 | Covid-19 Synthetic Minigene Vaccine |
| 2 | Tocilizumab | 2 | “Gam-COVID-Vac ", drug solution for intramuscular administration |
| 2 | Hydroxychloroquine + Lopinavir/Ritonavir+Interferon-β 1a/Interferon-β 1b | 2 | Lipid nanoparticle encapsulated mRNA-based vaccine (mRNA-1273) |
| 2 | Methylprednisolone | 2 | Lyofilizate drug “Gam-COVID-Vac Lyo" |
| 1 | Ivermectin | 2 | Heat-inactivated plasma from donors with COVID-19 (V-SARS) |
| 1 | Favipiravir | 2 | Vaccine with autologous dendritic cells loaded with antigens from SARS-CoV-2 (AV-COVID-19) |
| 2 | RNA vaccine candidate (BNT162b1) | ||
| 1 | Artificial antigen presenting cells vaccine | ||
| 1 | Recombinant SARS-CoV-2 Trimeric S Protein Subunit Vaccine | ||
| 1 | SARS-CoV-2 rS nanoparticle vaccine with or without Matrix-M | ||
| 1 | Bacterial medium with live bifidobacterium longum (bacTRL-spike) | ||
| 1 | Covid-19 Synthetic minigene vaccine | ||
| 1 | Candidate adjuvanted recombinant protein SARS-COV-2 vaccine (Covax-19™) | ||
| 1 | COVID19 vaccine |
*Trial phase.
9. Future development
Current vaccines/antiviral drugs are targeting 22 viruses; however, several emerging infectious diseases without any treatments are continuously affecting large populations. Henceforth, viruses such as Epstein-Barr virus (EBV), human norovirus, human rhinovirus, human herpesvirus 6, including currently pandemic human coronavirus, Zika virus, etc have gained a lot of attention (Clercq and Li, 2016). Recent efforts have been laid to find inhibitors for the recent Ebola virus during 2013–2016 for filoviruses. The nucleotide analog GS-5734, as a polymerase inhibitor is in clinical trials (Chaudhuri et al., 2018). For dengue infection, modipafant and celgosivir are in clinical trials. Modipavant activates the platelet-activating factor receptor (PAFR). Celgosivir, derived from a natural product inhibits alpha-glucosidase I (Chaudhuri et al., 2018). Brincidofovir is in a clinical trial for smallpox and adenovirus infection. Tecovirimat, which was identified by high throughput screening is also being developed for smallpox and inhibits p37, a viral protein (Chaudhuri et al., 2018). For influenza treatment, Japan has approved favipiravir under certain conditions however it has not been approved elsewhere. Baloxavir marboxil and pimodivir targeting the polymerase side of the influenza virus are in clinical trials from 2018 in the USA. These two compounds do not allow a virus to control block the transcription process in a virus (Chaudhuri et al., 2018).
The quest for new, improved antivirals for the influenza virus is still high though there are still vaccines against the virus. Out of 138 virus species, 59 of the viruses were found to be infectious for human beings from companion animals and were found to have zoonotic potential. Wild animal species were found to be the main source of new emerging viruses, leading to new diseases and epidemics (Reperant et al., 2016). To protect humans from harmful viruses, it is important to first identify methods that can be adopted to combat emerging viruses. Viruses are a potential threat to the world as they disperse easily and are communicable within days. Hence, a global surveillance system is necessary to combat these viruses. Proper investigation in the sources of new human viruses is also required (Woolhouse et al., 2012). The emergence of new viral infections and resistance to current antiviral therapy has urged scientists to design new antiviral agents. These antivirals can be divided into two groups: one that inhibits virus, and the other that inhibits host cells. Most antivirals target viral polymerases and proteases, while drug-resistant viruses are one of the main challenges in viral therapy. Another obstacle is that most of the viruses are eliminated but genetic material is left behind in the host, which can still cause the infection (Lou et al., 2014). The resistance of viruses to various drugs can only be overcome by designing antivirals against the conserved regions of the viruses (Saxena et al., 2010). Nevertheless, several antiviral inhibitors (brivudine, acyclovir, TDF, foscarnet, famciclovir, lamivudine, etc.) are used for the treatment of more than one virus. Perhaps more defined computational studies can be used in the future to design such compounds that can be used to target many infectious viruses.
In all kinds of infections, cytokine storms are apparent therefore a combination therapy having antiviral agents with immunotherapy may have significant effects (D́elia et al., 2013). It has been established now that SARS-CoV-2 induces strong cytokine storms which result in SARS-CoV-2 induced pneumonia. Hence, it is necessary to identify current existing therapies to treat hyper inflammation to reduce COVI-19 associated mortality (Nile et al., 2020). It has been reported that therapies of COVID 19 with glucocorticoids, chloroquine, tocilizumab have proved to be effective in COVID 19 patients. Moreover, it has been suggested that TNF blockers, IFN-αβ receptor blockers or antagonists ulinastatin, siponimod, eritoran, and other oxidized phospholipids inhibitors may also be proved to be effective against HCoV-induced inflammatory response. However, clinical trials and further investigation is necessary to ascertain their role in it (Ye et al., 2020). Given in Table 4 is the list of the drugs that have been completed at various clinical phases and the list of the vaccines that are in the various clinical phases. Most of the work in the clinical trial of vaccines work in clinical trials are on BCG and attenuated COVID 19 virus (ClinicalTrial.gov, 2020).
10. Conclusions and future perspectives
Throughout the last 50 years, enormous efforts have been made in the field of antiviral drug discovery, and many antiviral drugs are available to treat different human infectious diseases. To the best of our knowledge, this review presents, for the first time, a comprehensive overview of viruses discovered over the past 100 years. To emphasize the most recent development in antiviral drug discovery, the present studies highlight an absolute summary of potential antiviral agents. It is still a challenge for the scientific community to rapidly develop antiviral drugs that will continue to be essential for antiviral treatments against current and emerging viral infections. We anticipate that novel antiviral therapies, planning, and management might contribute towards the elimination and eradication of human infectious diseases in the future. We believe that new antivirals can be in the future to be approved and categorized for the cure of a broad range of emerging infectious diseases. In the end, it is also recommended that there is a need to increase societal awareness of high-risk animals that have a history of causing epidemics. Three-dimensional structure comparisons and sequence analysis of the protease structures of the three viruses–MERS, SARS, and SARS-CoV-2 have shown that perhaps in the future, a drug or vaccine can be formulated that may be effective against all three viruses.
Declaration of competing interest
No potential conflict of interest relevant to this article was reported.
References
- Abdel-Hakeem M.S. Viruses teaching immunology: role of LCMV model and human viral infections in immunological discoveries. Viruses 11. 2019 doi: 10.3390/v11020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adem S., Eyupoglu V., Sarfraz I., Rasul A., Ali M. Identification of potent COVID-19 main protease (Mpro) inhibitors from natural polyphenols: an in silico strategy unveils a hope against CORONA. Preprints 2020030333. 2020. [DOI] [PMC free article] [PubMed]
- Al Awaidy S.T., Khamis F. Middle East respiratory syndrome coronavirus (MERS-CoV) in Oman: current situation and going forward. Oman Med. J. 2019;34:181–183. doi: 10.5001/omj.2019.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander D.J. Newcastle disease and other avian paramyxoviruses. Rev. Sci. Tech. 2000;19:443–462. doi: 10.20506/rst.19.2.1231. [DOI] [PubMed] [Google Scholar]
- Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A.B., Flach B., Lin B.C., Doria-Rose N.A., O'Dell S., Schmidt S.D., Corbett K.S., Swanson P.A., Padilla M., Neuzil K.M., Bennett H., Leav B., Makowski M., Albert J., Cross K., Edara V.V., Floyd K., Suthar M.S., Martinez D.R., Baric R., Buchanan W., Luke C.J., Phadke V.K., Rostad C.A., Ledgerwood J.E., Graham B.S., Beigel J.H. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. NEJMoa2028436. 2020. [DOI] [PMC free article] [PubMed]
- Arulanandam C.D. Molecular docking study on the structure of COVID-19 main protease (MPro) to find the best viral inhibitor. figshare. Dataset. 2020. [DOI]
- Báez-Santos Y.M., St John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bkhaitan M.M. Simultaneous determination of simvastatin with caffeine in bulk drug, formulation and their monitoring in mice plasma through HPLC-PDA technique. Curr. Anal. Chem. 2017;13:1–7. doi: 10.2174/1573411013666170721112505. [DOI] [Google Scholar]
- Bosch A., Pintó R.M., Guix S. Human astroviruses. Clin. Microbiol. Rev. 2014;27:1048–1074. doi: 10.1128/CMR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenda L.T. 2020. Parainfluenza Virus Infections. MSD Manual. [Google Scholar]
- Bunney P., Zink A.N., Holm A., Billington C.J., Kotz C.M. Orexin activation counteracts decreases in nonexercise activity thermogenesis NEAT caused by high fat diet. Physiol. Behav. 2017;176:139–148. doi: 10.1016/j.physbeh.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprari S., Metzler S., Lengauer T., Kalinina O.V. Sequence and structure analysis of distantly-related viruses reveals extensive gene transfer between viruses and hosts and among viruses. Viruses. 2015;7:5388–5409. doi: 10.3390/v7102882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Napoli R. Di. 2020. Features, evaluation and treatment coronavirus (COVID-19). StatPearls publishing, treasure island. [PubMed] [Google Scholar]
- Chan J.F., Yuan S., Kok K., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., Tsoi H., Lo S.K., Chan K., Poon V.K., Chan W., Ip J.D. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission : a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S., Symons J.A., Deval J. Innovation and trends in the development and approval of antiviral medicines : 1987-2017 and beyond Innovation and trends in the development and approval of antiviral medicines : 1987 – 2017 and beyond. Antivir. Res. 2018;155:76–88. doi: 10.1016/j.antiviral.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Guo J., Wang C., Luo F., Yu X., Zhang W., Li J., Zhao D., Xu D., Gong Q., Liao J., Yang H. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet. 2020;395:809–815. doi: 10.1016/S0140-6736(20)30360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho A., Saunders O.L., Butler T., Zhang L., Xu J., Vela J.E., Feng J.Y., Ray A.S., Kim C.U. Bioorganic & medicinal chemistry letters synthesis and antiviral activity of a series of 1 0 -substituted. Bioorg. Med. Chem. Lett. 2012;22:2705–2707. doi: 10.1016/j.bmcl.2012.02.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choppin P.W., Compans R.W. Reproduction of paramyxoviruses. Comprehensive Virology. Springer Link. 1975:95–178. [Google Scholar]
- Choudhary A., Dar L. In: Bench to Bedside Diagnostic Microbiology for the Clinicians. Khardori N., editor. Taylor & Francis CRC Press; 2018. Microbiological Diagnosis of Viral Diseases. [Google Scholar]
- Chua K.B., Crameri G., Hyatt A., Yu M., Tompang M.R., Rosli J., McEachern J., Crameri S., Kumarasamy V., Eaton B.T., Wang L.F. A previously unknown reovirus of bat origin is associated with an acute respiratory disease in humans. Proc. Natl. Acad. Sci. Unit. States Am. 2007;104:11424–11429. doi: 10.1073/pnas.0701372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clercq E., Li D.C. Approved antiviral drugs over the past 50 years. Clin. Microbiol. Rev. 2016;29:695–747. doi: 10.1128/CMR.00102-15.Address. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ClinicalTrialgov [WWW Document] 2020. U.S. Natl. Libr. Med. [Google Scholar]
- Corstjens P.L.A.M., Abrams W.R., Malamud D. Saliva and viral infections. Periodontol. 2016;2000(70):93–110. doi: 10.1111/prd.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A.J. Herpesvirus systematics. Vet. Microbiol. 2010;143:52–69. doi: 10.1016/j.vetmic.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delia R.V., Harrison K., Oyston P.C., Lukaszewski R.A., Clark G.C. Targeting the “cytokine storm” for therapeutic benefit. Clin. Vaccine Immunol. 2013;20:319–327. doi: 10.1128/CVI.00636-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinter Z., Hermodsson S., Hermodsson L. Studies on Yucaipa: its classification as a member of the paramyxovirus group. Virology. 1964;22:297. doi: 10.1016/0042-6822(64)90020-0. [DOI] [PubMed] [Google Scholar]
- Douglas G., Samant V.B. 2013. The vaccine industry, in: Vaccines. [Google Scholar]
- Estrada-Peña A., De La Fuente J. The ecology of ticks and epidemiology of tick-borne viral diseases. Antivir. Res. 2014;108:104–128. doi: 10.1016/j.antiviral.2014.05.016. [DOI] [PubMed] [Google Scholar]
- Fauquet C.M., Mayo M.A., Maniloff J., Desselberger U., Ball L.A. In: Press E., editor. Eighth Report of the International Committee on Taxonomy of Viruses; London: 2005. Virus Taxonomy. [Google Scholar]
- Forni D., R C M.C., Sironi M. Molecular evolution of human Coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert T.W., Peter B.J. Exotic emerging viral diseases: progress and challenges. Nat. Med. 2004;10:S110–S121. doi: 10.1038/nm1142. [DOI] [PubMed] [Google Scholar]
- Goldenthal K.L., Midthun K., Zoon K.C. In: Medical Microbiology. Baron S., editor. University of Texas Medical Branch at Galveston; Galveston (TX): 1996. Control of Viral Infections and Diseases. [PubMed] [Google Scholar]
- Goo L., Dowd K.A., Lin T.Y., Mascola J.R., Graham B.S., Ledgerwood J.E., Pierson T.C. A virus-like particle vaccine elicits broad neutralizing antibody responses in humans to all chikungunya virus genotypes. J. Infect. Dis. 2016;214:1487–1491. doi: 10.1093/infdis/jiw431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner J. Three emerging coronaviruses in two decades: the story of SARS, MERS, and now COVID-19. Am. J. Clin. Pathol. 2020;153:420–421. doi: 10.1093/ajcp/aqaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Cao Q., Hong Z., Tan Y., Chen S., Jin H., Tan K., Wang D., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 ( COVID-19 ) outbreak – an update on the status. 2020;7:1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiou L., Zhou Y., Zhang M., Wang H., Zhao Q., Jing L. Updated approaches against SARS-CoV-2. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00483-20. e00483–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerbeck C.D., Whal Jensen V., Hooper J.W. In: Vaccines for Biodefense and Emerging and Neglected Diseases. Barrett A.D.T., Stanberry L.R., editors. Academic Press; 2009. Hantavirus. [Google Scholar]
- He J., Hu L., Huang X., Wang C., Zhang Z., Wang Y., Zhang D., Ye W. Potential of coronavirus 3C-like protease inhibitors for the development of new anti-SARS-CoV-2 drugs: insights from structures of protease and inhibitors. Int. J. Antimicrob. Agents. 2020;56(106055) doi: 10.1016/j.ijantimicag.2020.106055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrickson K.J. Parainfluenza Viruses. 2003;16:242–264. doi: 10.1128/CMR.16.2.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E.C., Rambaut A. Viral evolution and the emergence of SARS coronavirus 1059–1065. 2004. [DOI] [PMC free article] [PubMed]
- Huang Y. In: Preparing for the Next Disease Outbreak. Learning from S.A.R.S., editor. Workshop Summary. National Academies Press (US); Washington DC: 2004. The SARS epidemic and its aftermath IN China: a political perspective. [Google Scholar]
- Huang C., Wei P., Fan K., Liu Y., Lai L. 3C-like proteinase from SARS coronavirus catalyzes substrate hydrolysis by a general base mechanism. Biochemistry. 2004;43:4568–4574. doi: 10.1021/bi036022q. [DOI] [PubMed] [Google Scholar]
- Idda M.L., Soru D., Floris M. Overview of the first 6 Months of clinical trials for COVID-19 pharmacotherapy: the most studied drugs. Front. Publ. Health. 2020;8:1–7. doi: 10.3389/fpubh.2020.00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H.S., Jeong A., Cheon D.S. Epidemiology of astrovirus infection in children. Korean J. An. Pediatr. 2012;55:77–82. doi: 10.3345/kjp.2012.55.3.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S., Kim S., Shin D.H., Kim M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme Inhib. Med. Chem. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph A. 2020. As the corona virus spreads, a drug that once raised the world's hopes is given a second shot. Statnews. [Google Scholar]
- Khaerunnisa S., Kurniawan H., Awaluddin R., Suhartati S. March 2020. 2020. Potential inhibitor of COVID-19 main protease ( M pro ) from several medicinal plant compounds by molecular docking study. Prepr. | NOT PEER-REVIEWED | posted 13; pp. 1–14. [DOI] [Google Scholar]
- Khan M.F. Identification of dietary molecules as therapeutic agents to combat COVID-19 using molecular docking studies 1–17. 2020. [DOI]
- King A., Adams M., Carstens E., Lefkowitz E. Virus taxonomy: classification and nomenclature of viruses. Ninth rep. Int. Comm. Taxon. Viruses. 2012:193–210. ISBN 978-0-12-384684-6. [Google Scholar]
- Kirchmair J., Distinto S., Roman Liedl K., Markt P., Maria Rollinger J., Schuster D., Maria Spitzer G., Wolber G. Development of anti-viral agents using molecular modeling and virtual screening techniques. Infect. Disord. - Drug Targets. 2012;11:64–93. doi: 10.2174/187152611794407782. [DOI] [PubMed] [Google Scholar]
- Kneller D.W., Phillips G., O'Neill H.M., Jedrzejczak R., Stols L., Langan P., Joachimiak A., Coates L., Kovalevsky A. Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 2020;11:7–12. doi: 10.1038/s41467-020-16954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M., Chang S.Y., Byun S.Y., Choi I., Anne-Laure Pham Hung d’Alexandry d’Orengiani, Min J.-Y.D.S., Windisch M.P. Screening of FDA-approved drugs using a MERS-CoV clinical isolate from South Korea identifies potential therapeutic options for COVID-19 Meehyun Ko. bioRxiv Prepr. 2020 doi: 10.1101/2020.02.25.965582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl C., Lesnik R., Brinkmann A., Ebinger A., Radonić A., Nitsche A., Mühldorfer K., Wibbelt G., Kurth A. Isolation and characterization of three mammalian orthoreoviruses from European bats. PLoS One. 2012;7(8):e43106. doi: 10.1371/journal.pone.0043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug R.M. Clues to the virulence of H5N1 viruses in humans. Science (80-. ) 2006;311:1562–1563. doi: 10.1126/science.1125998. [DOI] [PubMed] [Google Scholar]
- Kumar S., Maurya V.K., Prasad A.K., Bhatt M.L.B., Saxena S.K. Structural, glycosylation and antigenic variation between 2019 novel Structural, glycosylation and antigenic variation between 2019 novel coronavirus ( 2019-nCoV ) and SARS coronavirus. VirusDisease. 2020;31:13–21. doi: 10.1007/s13337-020-00571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R.A., Parks G.D. sixth ed. 2012. Paramyxoviridae. Fields Virol. [DOI] [Google Scholar]
- Law G.L., Tisoncik-Go J., Korth M.J., Katze M.G. Drug repurposing: a better approach for infectious disease drug discovery? Curr. Opin. Immunol. 2013;25:588–592. doi: 10.1016/j.coi.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P.-H. Characterization and inhibition of SARS-coronavirus main protease. Curr. Top. Med. Chem. 2006;6:361–376. doi: 10.2174/156802606776287090. [DOI] [PubMed] [Google Scholar]
- Liu Z., Guo Z., Wang G., Zhang D., He H., Li G., Liu Y., Higgins D., Walsh A., Shanahan-Prendergast L., Lu J. Evaluation of the efficacy and safety of a statin/caffeine combination against H5N1, H3N2 and H1N1 virus infection in BALB/c mice. Eur. J. Pharmaceut. Sci. 2009;38:215–223. doi: 10.1016/j.ejps.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Live Science Staff Treatments for COVID-19: Drugs Being Tested against the Coronavirus. Livest. Sci. 2020 [Google Scholar]
- Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Pharm D., Gerber S.I., Kim L., Tong S., Ph D., Lu X., Lindstrom S., Ph D., Pallansch M.A., Ph D., Weldon W.C., Ph D., Biggs H.M., Uyeki T.M., Pillai S.K. First case of 2019 novel coronavirus in the United States. 2020. 929-936. [DOI] [PMC free article] [PubMed]
- Lou Z., Sun Y., Rao Z. Current progress in antiviral strategies. Trends Pharmacol. Sci. 2014;35:86–102. doi: 10.1016/j.tips.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma R.C.W., Holt R.I.G. COVID-19 and diabetes. Diabet. Med. 2020;3:1–3. doi: 10.1111/dme.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclachlan N.J., Edward J., D. fourth ed. 2010. Fenner’s Veterinary Virology. ISBN: 9780123751584. [Google Scholar]
- Marhöfer R.J., Oellien F., Selzer P.M. Drug discovery and the use of computational approaches for infectious diseases. Future Med. Chem. 2011;3:1011–1025. doi: 10.4155/fmc.11.60. [DOI] [PubMed] [Google Scholar]
- Mathew S., ThaniAl A.A., Yassine H.M. Computational screening of known broad-spectrum antiviral small organic molecules for potential influenza HA stem inhibitors. PLoS One. 2018;13(9) doi: 10.1371/journal.pone.0203148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCOLL K.A., N T., AGUILAR SETIEN A. Bat lyssavirus infections. Rev. Sci. Tech. 2000;19:177–196. doi: 10.20506/rst.19.1.1221. [DOI] [PubMed] [Google Scholar]
- MCKEEVER A. Dozens of COVID-19 vaccines are in development. Here are the ones to follow. Natl. NOW Times. 2020 (Geoghraphic) [Google Scholar]
- Mir M. Hantaviruses. Clin. Lab. Med. 2010;30:67–91. doi: 10.1038/jid.2014.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza A.Z. Advancement in the development of heterocyclic nucleosides for the treatment of cancer - a review. Nucleosides. Nucleotides and Nucleic Acids. 2019;38:836–857. doi: 10.1080/15257770.2019.1615623. [DOI] [PubMed] [Google Scholar]
- Muramatsu T., Takemoto C., Kim Y.T., Wang H., Nishii W., Terada T., Shirouzu M., Yokoyama S. SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity. Proc. Natl. Acad. Sci. U. S. A. 2016;113:12997–13002. doi: 10.1073/pnas.1601327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik P., Dave V.P., Id J.J. Detection of Torque Teno virus ( TTV ) and TTV- like minivirus in patients with presumed infectious endophthalmitis in India 1–8. 2020. [DOI] [PMC free article] [PubMed]
- Nichol S.T., Arikawa J., Kawaoka Y. From the Academy Emerging viral diseases. Proc. Natl. Acad. Sci. Unit. States Am. 2000;97:12411–12412. doi: 10.1073/pnas.210382297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nile S.H., Nile A., Qiu J., Li L., Jia X., Kai G. COVID-19: pathogenesis, cytokine storm and therapeutic potential of interferons. Cytokine Growth Factor Rev. 2020;53:66–70. doi: 10.1016/j.cytogfr.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhaus A. New respiratory viruses of humans. Pediatr. Infect. Dis. J. 27. 2008:S71–S74. doi: 10.1097/INF.0b013e3181684d7c. [DOI] [PubMed] [Google Scholar]
- Padron-Regalado E. Vaccines for SARS-CoV-2: lessons from other coronavirus strains. Infect. Dis. Ther. 2020;9:255–274. doi: 10.1007/s40121-020-00300-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F., Ye T., Sun P., Gui S., Liang B., Li L. Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology. 2020:1–15. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parija S.C., Thomas J.M. 2019. Human parainfluenza viruses (HPIV) and other parainfluenza viruses. MedScape. [Google Scholar]
- Parry W., Peterson E. 2020. Devastating infectious diseases. Live sci. [Google Scholar]
- Patel M.M., Hall A.J., Vinjé J., Parashar U.D. Noroviruses: a comprehensive review. J. Clin. Virol. 2009;44:1–8. doi: 10.1016/j.jcv.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Paul G., Menangle virus . Infectious Disease and Antimicrobial Agents. 2010. Pittsburgh. [Google Scholar]
- Pettersen E., Goddard T., Huang C., Couch G., Greenblatt D., Meng E., TE F. UCSF Chimera-A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.H. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J.R. Perspective piece mosquito-borne human viral Diseases : why Aedes aegypti ? Am. J. Trop. Med. Hyg. 2018;98:1563–1565. doi: 10.4269/ajtmh.17-0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard L.I., Chua K.B., Cummins D., Hyatt A., Crameri G.T., Wang L.-F.E.B. Pulau virus; a new member of the Nelson Bay orthoreovirus species isolated from fruit bats in Malaysia. Arch. Virol. 2006;151:229–239. doi: 10.1007/s00705-005-0644-4. [DOI] [PubMed] [Google Scholar]
- Proença-Módena J., Macedo I., Arruda E. H5N1 avian influenza virus: an overview. Braz. J. Infect. Dis. 2007;11:125–133. doi: 10.1590/s1413-86702007000100027. [DOI] [PubMed] [Google Scholar]
- Razonable R.R., Bobmphvft J.E.F., Tfsjof B.O.E., Joijcjupst Q. 2020. THERAPY ANTIVIRAL Antiviral Drugs for Viruses Other than Human Immunodeficiency Virus. [Google Scholar]
- Reperant L.A., Brown I.H., Haenen O.L., de Jong M.D., Osterhaus A.D.M.E., Papa A., Rimstad E., Valarcher J.F., Kuiken T. Companion animals as a source of viruses for human beings and food production animals. J. Comp. Pathol. 2016;155:S41–S53. doi: 10.1016/j.jcpa.2016.07.006. [DOI] [PubMed] [Google Scholar]
- RESEARCH HIGHLIGHT KAIMRC . 2019. The rapid journey of a deadly MERS outbreak [WWW Document]. KAIMRC Res. HIGHLIGHT. [Google Scholar]
- Riaz A. Recent understanding of the classification and life cycle of herpesviruses- A review. Sci. Lett. 2017;5:195–207. [Google Scholar]
- Robinson J. Everything you need to know about the COVID-19 therapy trials [WWW Document] Pharm. J. 2020 doi: 10.1211/PJ.2020.20208126. [DOI] [Google Scholar]
- Ryu W.-S. Molecular Virology of Human Pathogenic Viruses. Elsevier; 2017. Herpesviruses. [Google Scholar]
- Salim B., Noureddine M. Identification of compounds from nigella sativa as new potential inhibitors of 2019 novel coronasvirus (Covid-19): molecular docking study. ChemRxiv. 2020. [DOI]
- Saxena S.K., Saxena S., Saxena R., Arvinda M.L., Gupta A., Nair M.P.N. Emerging trends. Challenges and Prospects in Antiviral Therapeutics and Drug Development for Infectious Diseases. 2010;6:26–31. [Google Scholar]
- Shamshad H., Hafiz A., Althagafi I.I., Saeed M., Mirza A.Z. Characterization of the trypanosoma brucei pteridine reductase active- site using computational docking and virtual screening techniques. Curr. Comput. Aided Drug Des. 2020;16:583–598. doi: 10.2174/1573409915666190827163327. [DOI] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Leist S.R., Schäfer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Babusis D., Clarke M.O., Spahn J.E., Bauer L., Sellers S., Porter D., Feng J.Y., Cihlar T., Jordan R., Denison M.R., Baric R.S. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2012. [DOI] [PMC free article] [PubMed]
- Smith D.B., Becher P., Bukh J., Gould E.A., Meyers G., Monath T., Muerhoff A.S., Pletnev A., Rico-Hesse R., Stapleton J.T., Simmonds P. Proposed update to the taxonomy of the genera Hepacivirus and Pegivirus within the Flaviviridae family. J. Gen. Virol. 2016;97:2894–2907. doi: 10.1099/jgv.0.000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling J.C. Infections and Infestations. 2016. Viral infections; p. 5. [Google Scholar]
- Stollenwerk N., Harper R.W., Sandrock C.E. Bench-to-bedside review: rare and common viral infections in the intensive care unit - linking pathophysiology to clinical presentation. Crit. Care. 2008;12 doi: 10.1186/cc6917. Pubmed Partial articletitle stitle Volume Page. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J.H., Strauss E.G. 2008. Minus-strand RNA viruses, in: in viruses and human disease. [Google Scholar]
- Suthar M.S., Diamond M.S., Jr. West Nile virus infection and immunity. Nat. Rev. Microbiol. 2013;11:115–128. doi: 10.1038/nrmicro2950. [DOI] [PubMed] [Google Scholar]
- Tahamtan A., Inchley C.S., Marzban M., Tavakoli-yaraki M., Teymoori-rad M., Nakstad B. The role of microRNAs in respiratory viral infection. friend or foe. 2016:389–407. doi: 10.1002/rmv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahir M., Alqahtani S.M., Alamri M.A., Chen L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants†. J. Pharm. Anal. 2020;10(4):313–319. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Iwasa Y., Hijikata M., Mishiro S. Identification of a new human DNA virus (TTV-like mini virus, TLMV) intermediately related to TT virus and chicken anemia virus. Arch. Virol. 2000;145:979–993. doi: 10.1007/s007050050689. [DOI] [PubMed] [Google Scholar]
- Thom K., Morrison C., Lewis J.C.M., Simmonds P. Distribution of TT virus (TTV), TTV-like minivirus, and related viruses in humans and nonhuman primates. Virology. 2003;306:324–333. doi: 10.1016/s0042-6822(02)00049-1. [DOI] [PubMed] [Google Scholar]
- Tsunetsugu-yokota Y., Ohnishi K., Takemori T. 2006. Severe acute respiratory syndrome ( SARS ) coronavirus : application of monoclonal antibodies and development of an effective vaccine 117–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorg. Med. Chem. Lett. 2020;30:127377. doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dorn A. COVID-19 and readjusting clinical trials. Lancet (London, England) 2020;396:523–524. doi: 10.1016/S0140-6736(20)31787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walder R. Electron microscopic evidence of Nariva virus structure. J. Gen. Virol. 1971;123 doi: 10.1099/0022-1317-11-2-123. [DOI] [PubMed] [Google Scholar]
- Wang J. Fast identification of possible drug treatment of coronavirus disease -19 (COVID-19) through computational drug repurposing study. ChemRxiv. 2020. [DOI] [PMC free article] [PubMed]
- Wang L.F., Crameri G. Emerging zoonotic viral diseases. OIE Rev. Sci. Tech. 2014;33:569–581. doi: 10.20506/rst.33.2.2311. [DOI] [PubMed] [Google Scholar]
- Weaver S.C., Charlier C., Vasilakis N., Lecuit M. Zika, chikungunya, and other emerging vector-borne viral diseases. Annu. Rev. ofMedicine. 2018;69:395–408. doi: 10.1146/annurev-med-050715-105122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M., Scott F., Hudson Z., Howey R., Chase-Topping M. Human viruses: discovery and emeraence. Philos. Trans. R. Soc. B Biol. Sci. 2012;367:2864–2871. doi: 10.1098/rstb.2011.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Hua C., Wang Y., Wang Q., Xu Y., Li M., Mengzhu L., Xingzhou Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs bycomputational methods. Acta Pharm. Sin. B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P., Dong J., Zhao J., Wang F. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanping Z. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liuxingbingxue Zazhi. 2020;41:145–151. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003. [DOI] [PubMed] [Google Scholar]
- Yazdany J., Kim A.H.J. Use of hydroxychloroquine and chloroquine during the COVID-19 Pandemic : what every clinician should know. Ann. Intern. Med. 1–3. 2020. [DOI] [PMC free article] [PubMed]
- Ye Q., Wang B., Mao J. The pathogenesis and treatment of the ‘Cytokine Storm’’ in COVID-19.’ J. Infect. 2020;80:607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuill T.M. 2018. Overview of arbovirus, arenavirus, and filovirus infections. [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science (80-. ) 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Li H., Liang L.W., Y D H., Guo C. 2016. On the classification of lyssaviruses. J vet sci med 5. [Google Scholar]