Fig 3. Plasma metabolites altered by chemical class in lean healthy control subjects between ethnicities.
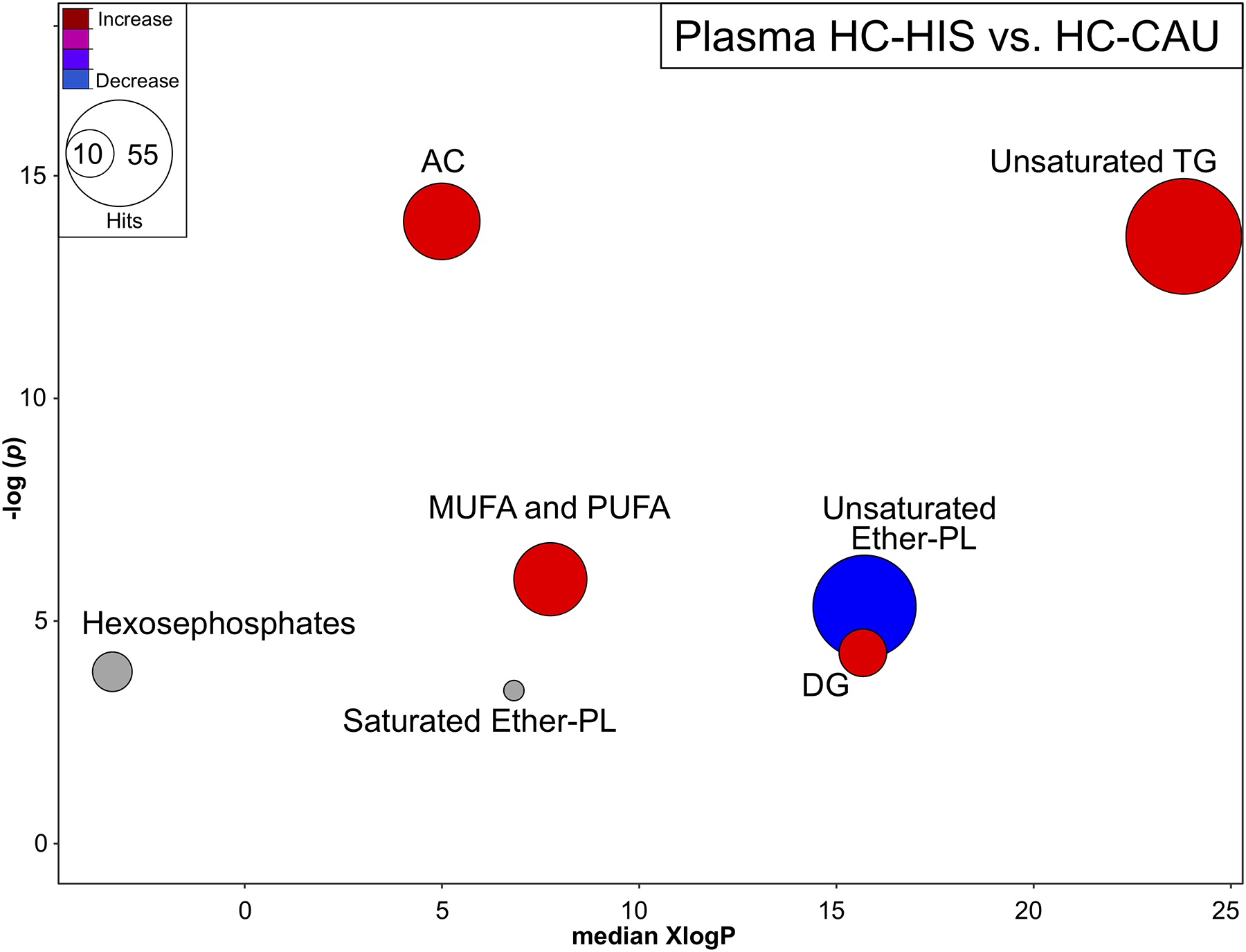
Chemical similarity enrichment analysis (ChemRICH) highlighting plasma metabolomic changes observed in HC-HIS, compared to HC-CAU. Each cluster represents altered chemical class of metabolites (p <0.05). Cluster sizes represent the total number of metabolites. Cluster color represents the directionality of metabolite differences: red – higher in HC-HIS; blue – lower in HC-HIS. Colors in between refer to mixed population of metabolites manifesting both higher and lower levels in HC-HIS when compared to the HC-CAU. The x-axis represents the cluster order on the chemical similarity tree. The plot y-axis shows chemical enrichment p-values calculated using Kolmogorov–Smirnov test. Only clusters with p <0.05 are shown. FDR-adjustment q =0.2, and clusters with FDR-adjusted p ≥0.2 are shown in gray. (n, HC-HIS =14, HC-CAU =8) The detailed ChemRICH results are shown in (Table S4). AC, Acylcarnitines; CAU, White Caucasian; DG, Diglycerides; Ether-PC, Ether-linked phospholipids; HIS, Hispanic; MUFA, Monounsaturated fatty acid; PUFA, Polyunsaturated fatty acids; TG, Triglycerides.
