Fig. 2: CcsBA transmembrane helices and its heme binding domains.
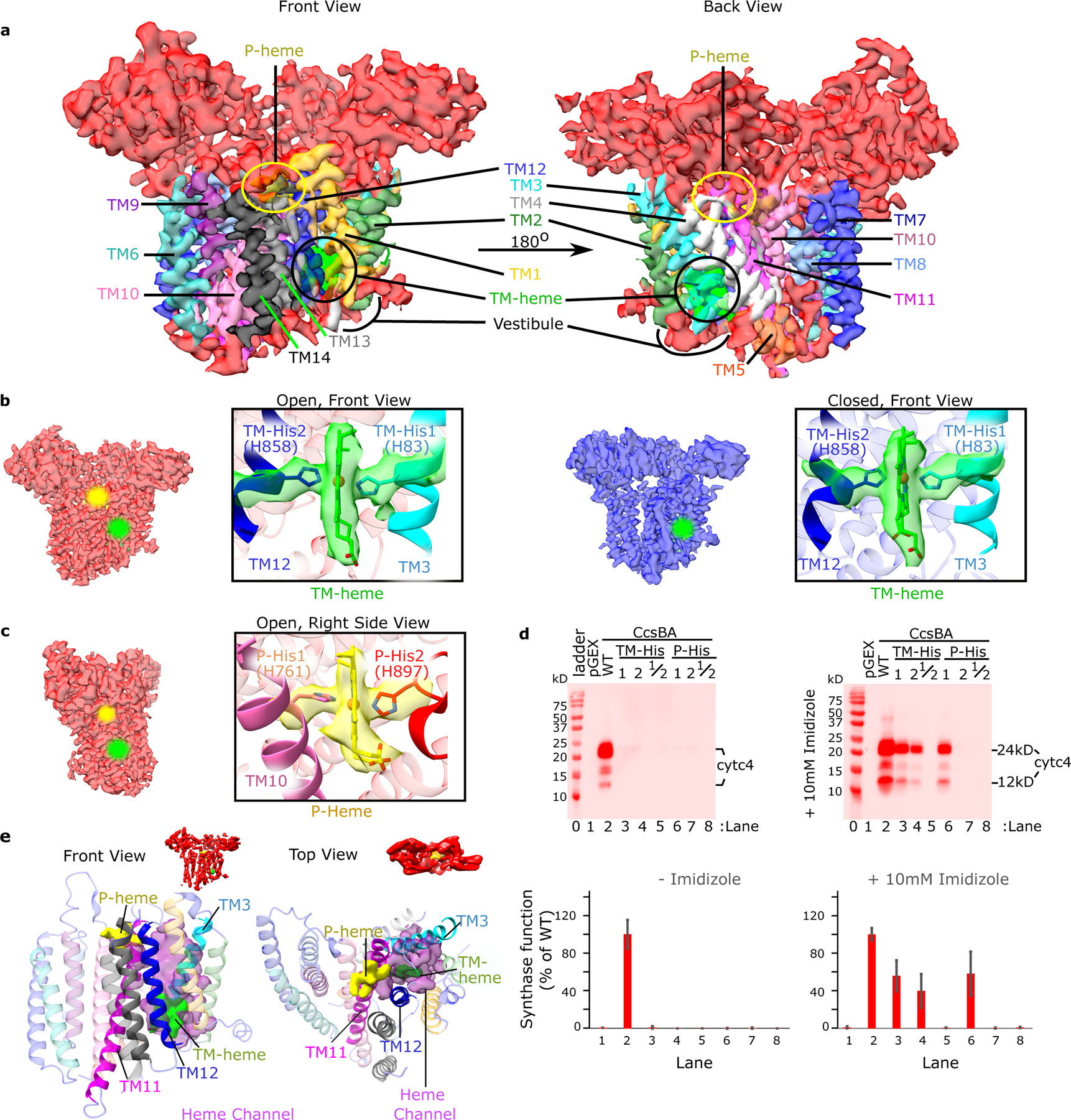
a. Front and back views of the open conformation of CcsBA cryo-EM map at 3.56 Å with labeled TMs, hemes, and the heme vestibule. b. CcsBA-open cryo-EM map (red) at 3.56 Å, next to the CcsBA-closed cryo-EM map (blue) at 4.14 Å. The boxed panels are a close-up view of the protein models docked into the electron density of the TM-heme (TMs 1 and 2 are hidden for better visualization of heme). c. A right side view rotated approximately 90° from that in b. The boxed panel is a close-up view of the protein-heme model of CcsBA-open docked into the electron density of P-Heme (TMs 8 and 9 are hidden to better visualize heme). To mark the heme locations, halos were added in the b and c cryo-EM densities. d. Representative heme stains of the reporter, cyt c4 in E. coli Δccm are shown and were used to assess the function of CcsBA His variants. Controls were pGEX vector (no CcsBA, lane 1) and wild-type CcsBA(WT, lane 2). Single and double His to Gly substitutions were made to produce variants at positions TM-His1, TM-His2, TM-His1/2, P-His1, P-His2, and P-His 1/2 that were nonfunctional. Results of chemical complementation of CcsBA His variants with 10mM imidazole (right panel). The bar graph (mean, n=3) and error bars (SD of the mean) are from representative data from 1 of 3 independent trials. e. For elucidation of the channel (magenta), P-heme (yellow) was modelled into the closed state based on the position of P-His 1 and the WWD domain. Then Pymol was used to find the channel cavity connecting TM-heme (green) to P-heme and exported to ChimeraX for display. The proposed channel lies between TM11 (magenta) and TM12 (blue).
