Extended Data Fig. 3. RNA-seq and ATAC-seq changes at Runx1, TCF1, and Ets1-bound genes.
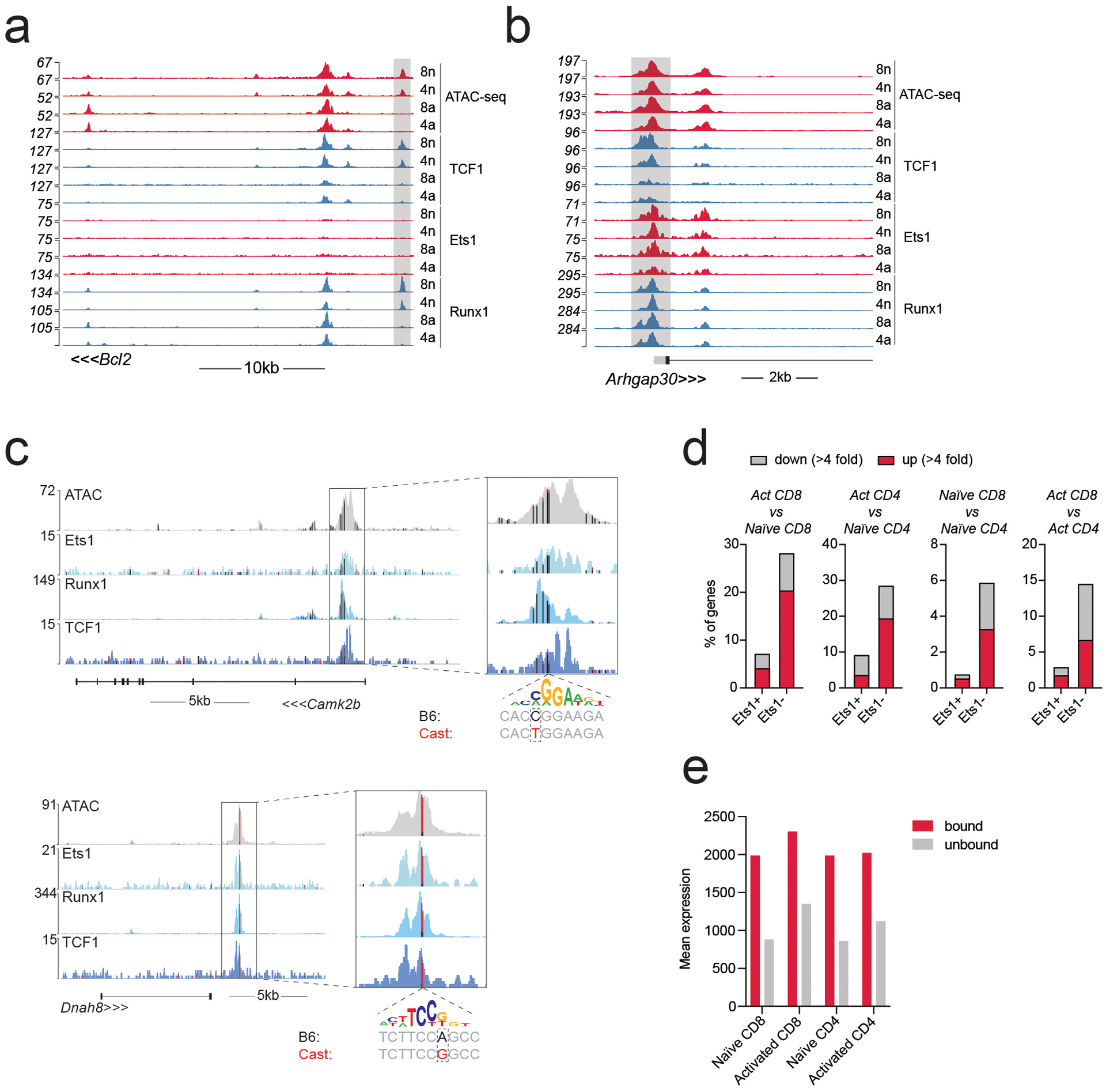
a-b: Gene tracks of RNA-seq, ATAC-seq and CUT&RUN coverage at the Bcl2 and Arhgap30 loci in naïve and activated CD8 T cells.
c: Track examples showing the effect of selected Ets1 motif variants on allelic bias in chromatin accessibility and TF binding in activated CD8 T cells from (B6/Cast) F1 mice. Vertical bars mark the position of genetic variants, with colors indicating the fraction of variant-containing reads coming from either the B6 (black) or Cast (red) allele. A genetic variant in the Camk2b locus results in a stronger Ets1 motif match on the B6 allele and a weaker match on the Cast allele and is associated with B6-specific chromatin accessibility and TF binding. The opposite pattern is observed at the Dnah8 locus.
d: Gene expression changes at genes (>100 reads in at least one cell type) nearest to Ets1-bound and -unbound ATAC-seq peaks.
e: Mean expression of genes nearest to Ets1-bound and -unbound ATAC-seq peaks.
