Figure 6: Deep whole RNA sequencing showed splicing defects in DDX41 deficient cells.
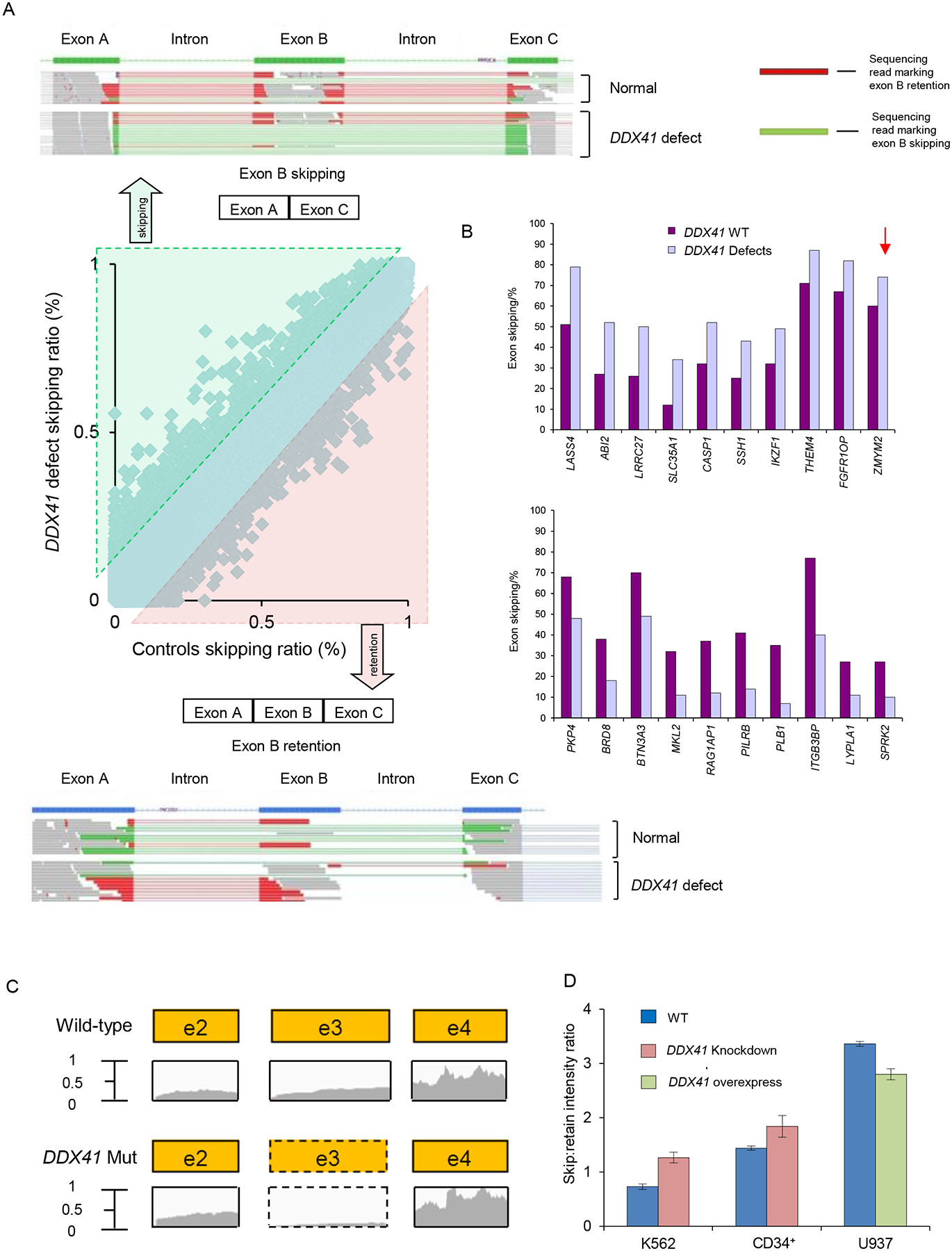
(A) Increased exon skipping (top) and retention (bottom) in patients with DDX41 defects are indicated by an excess of green reads and red reads, respectively. The center panel shows a scatter plot of exon skipping in RNA isolated from control cells versus RNA from DDX41 defective mutant cells. Lines show the 10% difference cutoff limit used to select the most frequently affected exons.
(B) Deep RNA sequencing was performed for blasts from patients with DDX41 mutations, deletions and wild-type to analyze altered splicing. The bar diagrams indicate the top 10 genes significantly more skipped in DDX41 defects (top) and in DDX41 wild-type (bottom). The arrow indicates the 13% difference of exon skipping in the ZMYM2 gene when comparing DDX41 defect and wild-type samples.
(C) Exon 3 of ZMYM2 was skipped in DDX41 deficient cells as demonstrated by the read counts from deep sequencing.
(D) RT-PCR was performed in K562 cells, CD34+ progenitors and U937 cells to evaluate ZMYM2 exon 3 skipping compared with controls. Depicted is the skip:retain intensity ratio for wild-type and DDX41 knockdown/overexpressing samples. Each bar represents the mean±SEM of 3 independent experiments.
