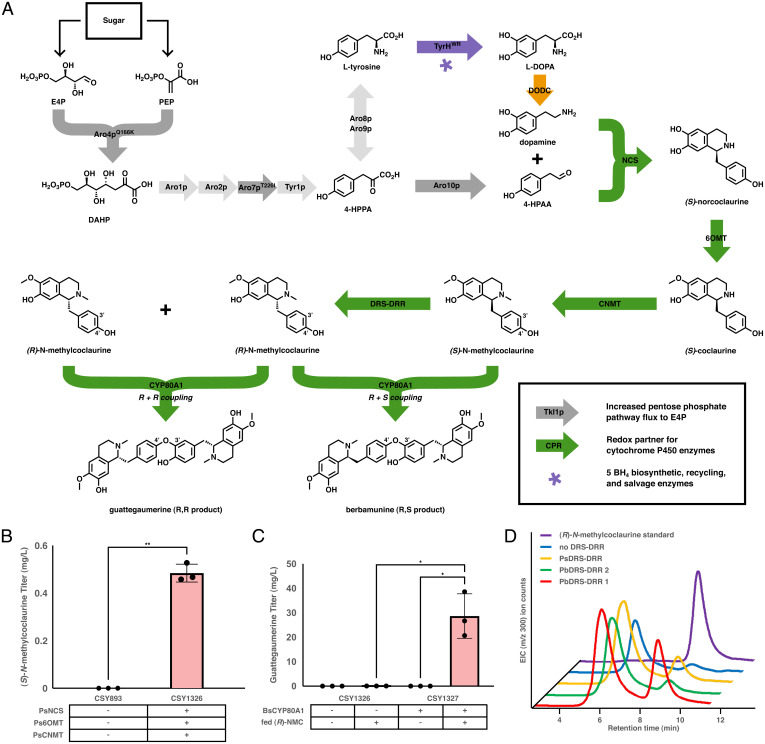
Fig. 1.
Biosynthetic pathway for de novo bisBIA biosynthesis. (A) Biosynthetic pathway from fed sugar to the bisBIA products berbamunine and guattegaumerine. The color of the arrows represents the species of origin of the enzyme employed: light gray, unmodified native yeast enzyme; dark gray, modified native yeast enzyme; green, enzyme of plant origin; orange, enzyme of bacterial origin; purple, enzyme of mammalian origin. (B) De novo production of (S)-N-methylcoclaurine in engineered yeast strains. Strains were cultured in synthetic complete media with 2% dextrose at 30 °C for 96 h before LC-MS/MS analysis of the growth media using the MRM transitions reported in SI Appendix, Table S5. (C) Production of the bisBIA guattegaumerine in a yeast strain expressing BsCYP80A1 and fed exogenous (R)-N-methylcoclaurine. Strains were cultured in selective media (YNB-Ura) with 2% dextrose at 30 °C for 96 h before being pelleted, resuspended in an equal volume of 200 µM (R)-NMC in 50 mM HEPES at pH 7.6, and incubated an additional 72 h at 30 °C before LC-MS/MS analysis of the media using the MRM transitions reported in SI Appendix, Table S5. (D) De novo production of (R)-N-methylcoclaurine in strain CSY1326 expressing PsCPR and different DRS-DRR enzymes off of low-copy plasmids. Strains were cultured in selective media (YNB-DO) with 2% dextrose at 30 °C for 96 h before extraction, purification, and analysis by normal-phase chiral chromatography. Data for B and C are presented as mean values ± the SD of three biologically independent samples. Asterisks represent Student’s two-tailed t test: *P < 0.05, **P < 0.01. Exact P values are given in SI Appendix, Table S6. Source data underlying B and C are provided in Dataset S1.