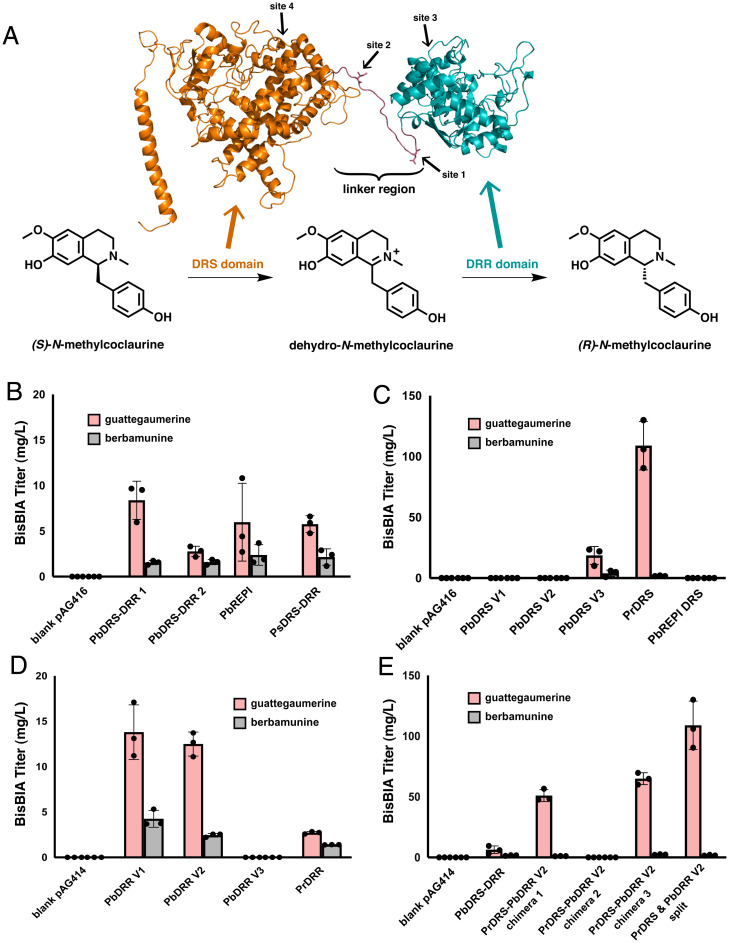
Fig. 3.
Engineering an optimized NMC epimerase for bisBIA production. (A) Homology model of PbDRS-DRR 1 and reaction schematic of the epimerization of (S)-N-methylcoclaurine to (R)-N-methylcoclaurine. The homology model was constructed with RaptorX (51) using CYP76AH1 from S. miltiorrhiza (44) as the basis of the DRS domain (orange) and 4,2’,4’,6’-tetrahydroxychalcone reductase from Medicago sativa (52) as the basis of the DRR domain (teal). By analogy to the reactions performed on the native substrate of PbDRS-DRR 1, (S)-reticuline, the DRS domain (orange) oxidizes (S)-N-methylcoclaurine to the intermediate dehydro-N-methylcoclaurine, which is then reduced by the DRR domain (teal) to the product (R)-N-methylcoclaurine. The four sites explored in C–E for cleaving the DRS and DRR domains of PbDRS-DRR 1 apart are indicated on the homology model with arrows. (B) De novo production of bisBIAs in yeast strains expressing natural DRS-DRR fusion proteins. Strains were cultured in selective media (YNB-Ura) with 2% dextrose with 2 mM L-DOPA and 10 mM ascorbic acid at 25 °C for 96 h before LC-MS/MS analysis of the growth media using the MRM transitions reported in SI Appendix, Table S5. (C) De novo production of bisBIAs in yeast strains expressing PbDRR V2 with different DRS domains as individual polypeptides. Strains were cultured in selective media (YNB-Ura-Trp) with 2% dextrose with 2 mM L-DOPA and 10 mM ascorbic acid at 25 °C for 96 h before LC-MS/MS analysis of the growth media using the MRM transitions reported in SI Appendix, Table S5. (D) De novo production of bisBIAs in yeast strains expressing PbDRS V3 with different DRR domains as individual polypeptides. Strains were cultured in selective media (YNB-Ura-Trp) with 2% dextrose with 2 mM L-DOPA and 10 mM ascorbic acid at 25 °C for 96 h before LC-MS/MS analysis of the growth media using the MRM transitions reported in SI Appendix, Table S5. (E) De novo production of bisBIAs in yeast strains expressing the two best performing DRS and DRR domains. The activity of the PrDRS and PbDRR V2 domains were tested as chimeric fusion proteins (sequences provided in SI Appendix, Table S7) and individual polypeptides. The native fused PbDRS-DRR 1 construct that was employed in previous figures is also shown for comparison. Strains were cultured in selective media (YNB-Ura for fusion proteins, YNB-Ura-Trp for split constructs) with 2% dextrose with 2 mM L-DOPA and 10 mM ascorbic acid at 25 °C for 96 h before LC-MS/MS analysis of the growth media using the MRM transitions reported in SI Appendix, Table S5. Data in B–E are presented as mean values ± the SD of three biologically independent samples. Source data underlying B–E are provided in Dataset S1.