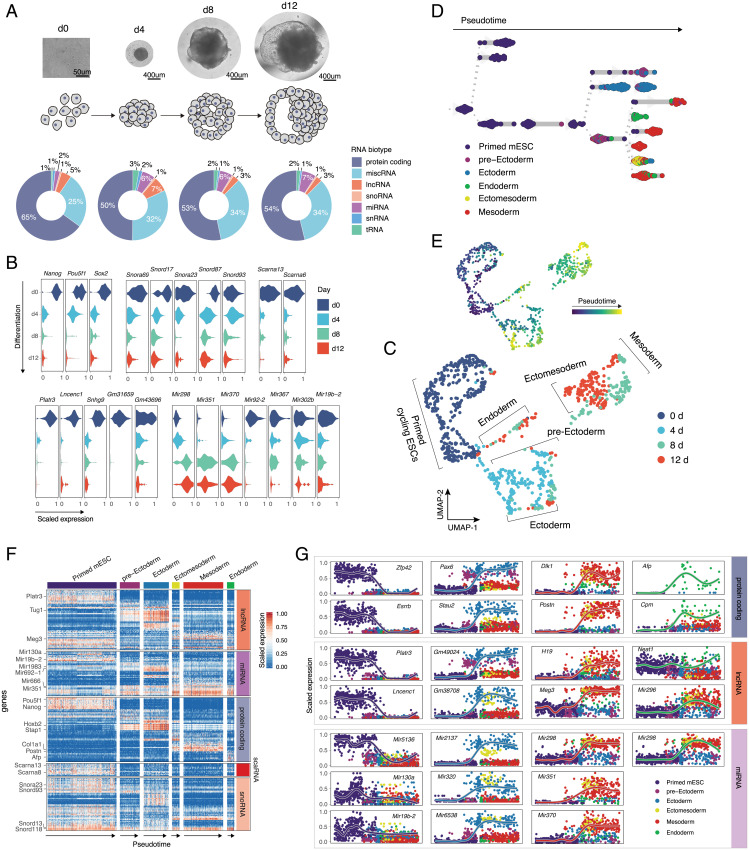
Fig. 3.
Coding and noncoding signature of differentiated single mESCs. (A) Microscope images and corresponding schematic representations of EB formation at four sampled time points. Pie charts represent distribution of mapped reads across RNA types. Genes were assigned to a specific biotype based on GENCODE M23 annotation for the reference chromosomes. tRNA was quantified by mapping the reads, nonmapping to any other RNA type, to the high-confidence gene set obtained from GtRNAdb. (B) Exemplary coding and noncoding genes that are up- or down-regulated during EB formation. Subpanels are grouped according to RNA type. (C) UMAP plot of collected cells colored by timepoint. Cells were clustered using a k nearest-neighbor algorithm and cell lineages were annotated based on the expression of marker genes within the identified clusters. (D) Lineage tree of EB differentiation. Each dot represents a cell colored according to the assigned lineage. Cells are arranged according to the computed pseudotime. (E) UMAP plot of collected cells colored by pseudotime. (F) Heatmap showing the variability in coding and noncoding gene expression across identified clusters. (G) Temporal and lineage-specific expression of selected protein-coding, lncRNA, and miRNA genes. Each column from left to right shows genes specific to: pluripotency state, ectoderm, mesoderm, or endoderm lineages.