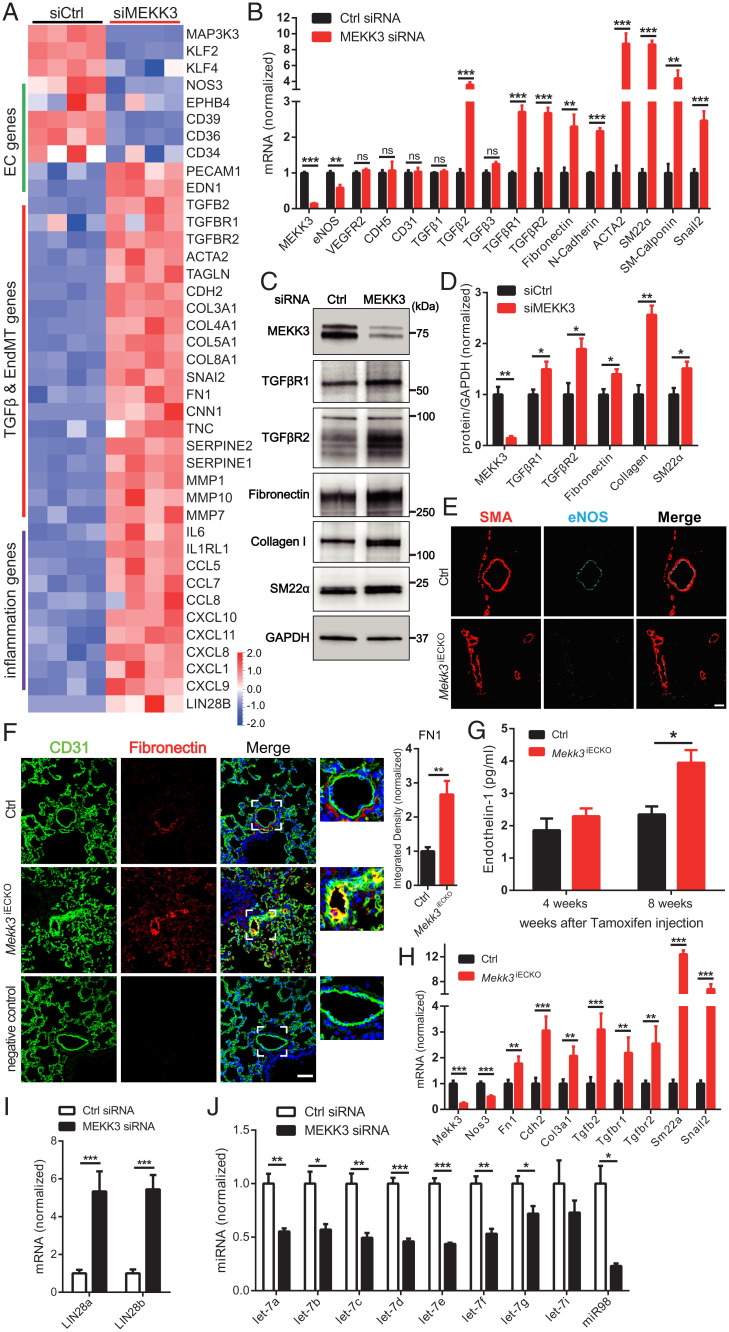
Fig. 3.
Loss of MEKK3 in ECs induces TGFβ-signaling. (A) Bulk RNA-seq analysis of HUVECs treated with Ctrl or MEKK3 siRNA. Heatmap shows EC, TGFβ, EndMT genes, and inflammatory genes. n = 4 samples for each group. (B) qpcr analysis of TGFβ pathway and EndMT markers expression in HUVECs treated with Ctrl or MEKK3 siRNA (n = 3). Data represent mean ± SD. (C and D) Representative WB and densitometric quantification of MEKK3 (n = 3), TGFβR1 (n = 4), TGFβR2 (n = 3), fibronectin (n = 3), collagen (n = 3), and SM22α (n = 3) expression in HUVECs treated with Ctrl or MEKK3 siRNA. Data represent mean ± SEM. (E) Immunostaining of eNOS and SMA in Ctrl and Mekk3iECKO lungs. (Scale bar, 20 μm.) (F) Immunostaining and quantification of Fibronectin in Ctrl and Mekk3iECKO lungs. Negative control means no primary antibody control. (Scale bar, 25 μm,) Data represent mean ± SEM. (G) Circulating endothelin-1 concentration in plasma from Ctrl and Mekk3iECKO mice at 4 and 8 wk after tamoxifen injection. n = 4 male mice per group. (H) qpcr analysis of EndMT markers in lung ECs isolated from Ctrl and Mekk3iECKO mice. Data represent mean ± SD. n = 4 mice per group. (I) qpcr analysis of LIN28a and LIN28b (n = 4) expression in HUVECs treated with Ctrl or MEKK3 siRNA. Data represent mean ± SD. (J) qpcr analysis of let-7 miRNA family (n = 3) expression in HUVECs treated with Ctrl or MEKK3 siRNA. ns: not significant, *P < 0.05, **P < 0.01, and ***P < 0.001, calculated by unpaired t test (B, D, F, H, I, and J) and two-way ANOVA with Sidak’s multiple comparison tests (G).