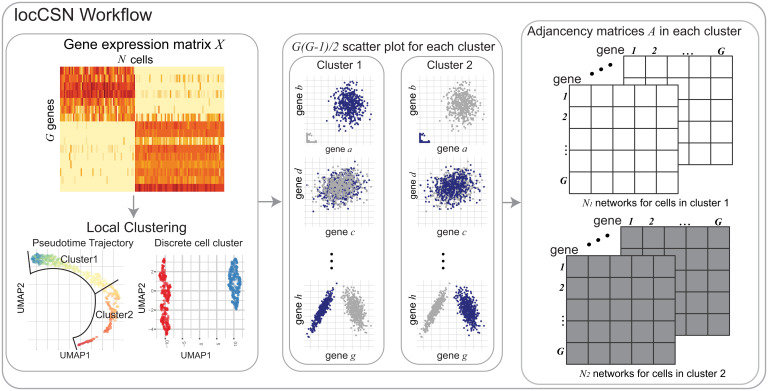
Fig. 1.
Workflow of locCSN. Starting with the gene expression matrix, we cluster homogeneous cells. For each cell in a cluster, we examine the coexpression of each pair of genes to determine whether the joint expression is unusually dense in a neighborhood of the cell, relative to expectation assuming the genes are independent; if so, the genes are connected in that cell’s network. For each cell, we then construct the gene–gene network, based on the results of the local independence test for each pair of genes.