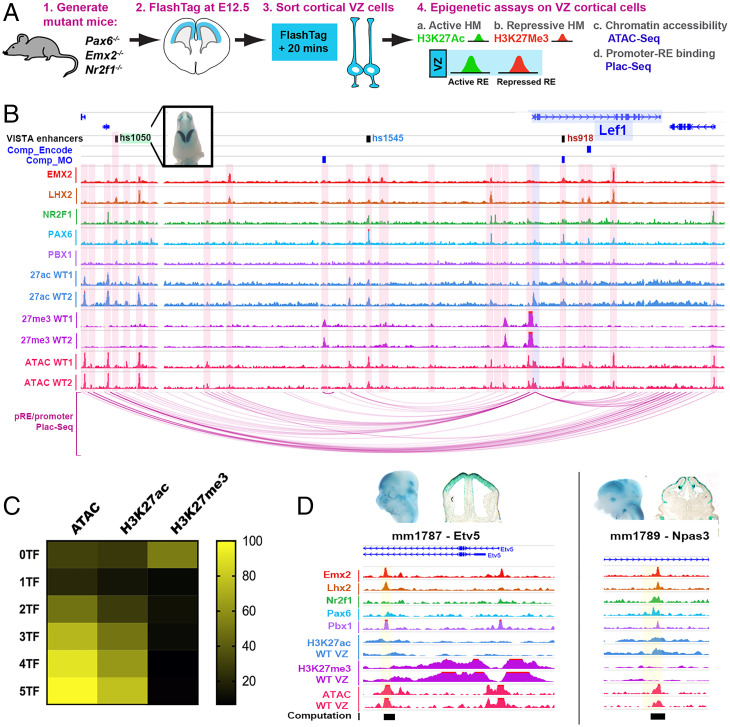
Fig. 6.
pRE landscape around CRTFN TF genes in cortical progenitors. (A) Method for VZ cell purification for epigenomic experiments (Materials and Methods). (B) Coverage of TF ChIPs, VZ histone marks (active: H3K27ac; repressive: H3K27me3), and VZ ATAC-seq in the Lef1 locus at E12.5. pRE–promoter interactions are derived from PLAC-seq (purple arcs Below) and computationally (blue boxes Above, Comp_Encode and Comp_MO; Dataset S5). VISTA enhancers are also indicated (green = cortical activity, blue = noncortical activity, and red = inactive). (C) Heatmap of percentage enrichment of epigenomic marks over pREs with differential combinatorial TF binding (0 to 5 TFs). (D) Enrichment of TF and epigenomic marks over newly identified cortical enhancers near CRTFN genes: mm1787 (Etv5) and mm1789 (Npas3).