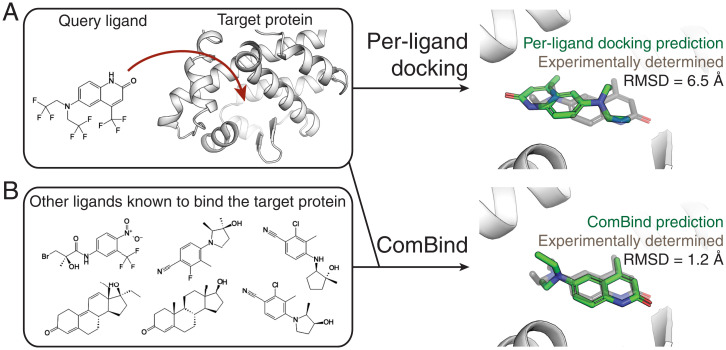
Fig. 1.
ComBind leverages nonstructural data to improve ligand binding pose predictions. (A) Standard docking methods take as input the chemical structure of the query ligand and the 3D structure of the target protein and predict a binding pose using a per-ligand scoring function. (B) ComBind additionally considers other ligands known to bind the target protein (whose binding poses are not known), resulting in more-accurate predictions. For clarity, hydrogen and fluorine atoms are omitted from the 3D renderings.