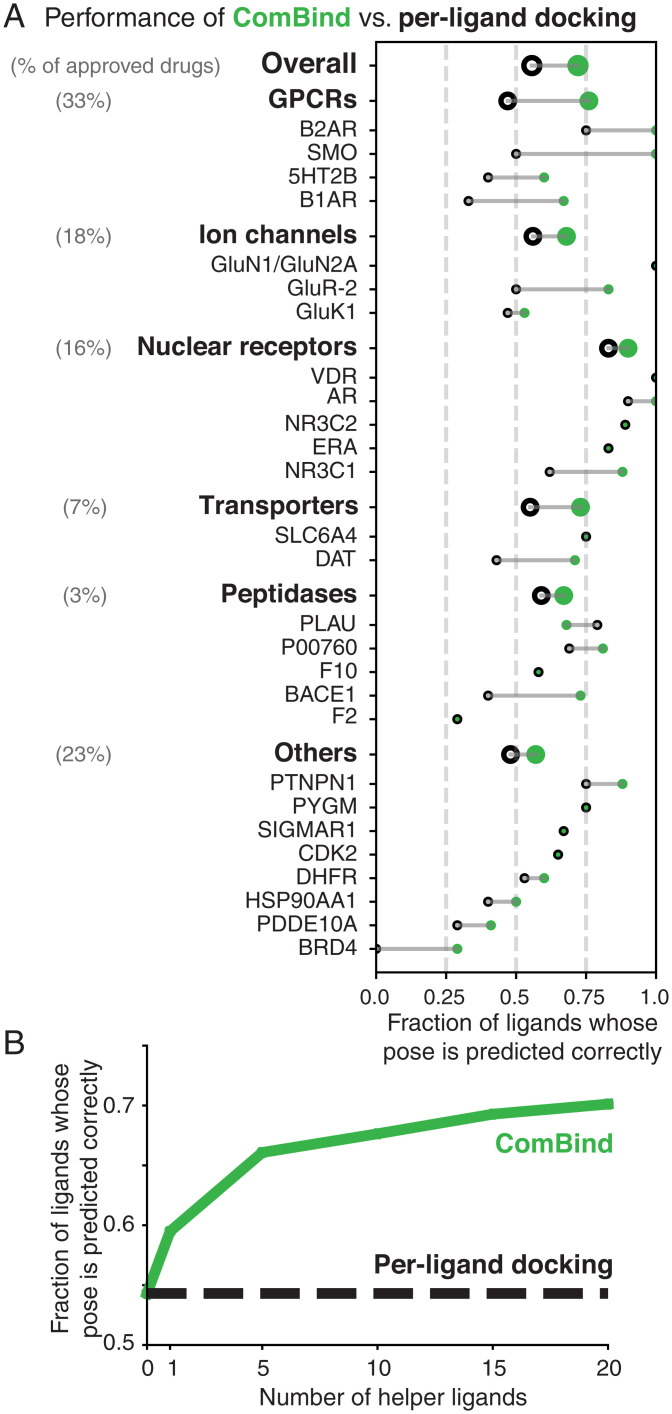
Fig. 4.
ComBind outperforms per-ligand docking on a diverse benchmark set. Performance of ComBind, as compared to a per-ligand scoring function, using helper ligands selected automatically from ChEMBL. All results are for “cross-docking” (the query ligand is docked into a structure determined in the presence of a distinct ligand). (A) Performance per target protein, target protein family (GPCRs, ion channels, etc.), and overall. Green disks and black circles indicate performance for ComBind and a state-of-the art per-ligand docking software package (Glide), respectively. (B) Performance as a function of the number of helper ligands. When using no helper ligands, ComBind is equivalent to per-ligand docking.