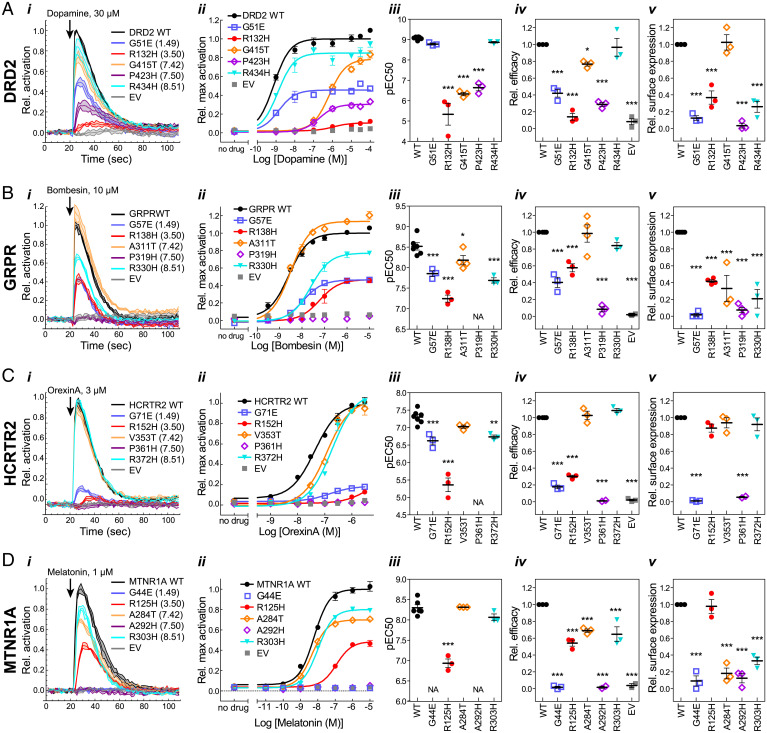
Fig. 4.
G protein activation by (A) DRD2, (B) GRPR, (C) HCRTR2, and (D) MTNR1A mutants. (i) Representative time courses showing G protein activation at a single ligand concentration. Lines indicate means ± SD of technical replicates at each timepoint. (ii) Representative dose–response curves generated from maximum activation values from data as in i. Points indicate means ± SEM of technical replicates. (iii–v) EC50 (iii) and efficacy (Emax) (iv) were determined from dose–response curve fits, and surface expression (v) was measured by cell ELISA, as described in Materials and Methods. Points represent means from independent experiments, and error bars show SEM. Mutants were compared to WT using one-way ANOVA and Dunnett’s posttest (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001). Mutants for which an EC50 could not be determined are indicated by “NA.” For mutants which did not reach a plateau in the dose–response curves, EC50 and efficacy determinations are estimates and represent lower limits. Error bars that are not visible are smaller than the associated symbol.