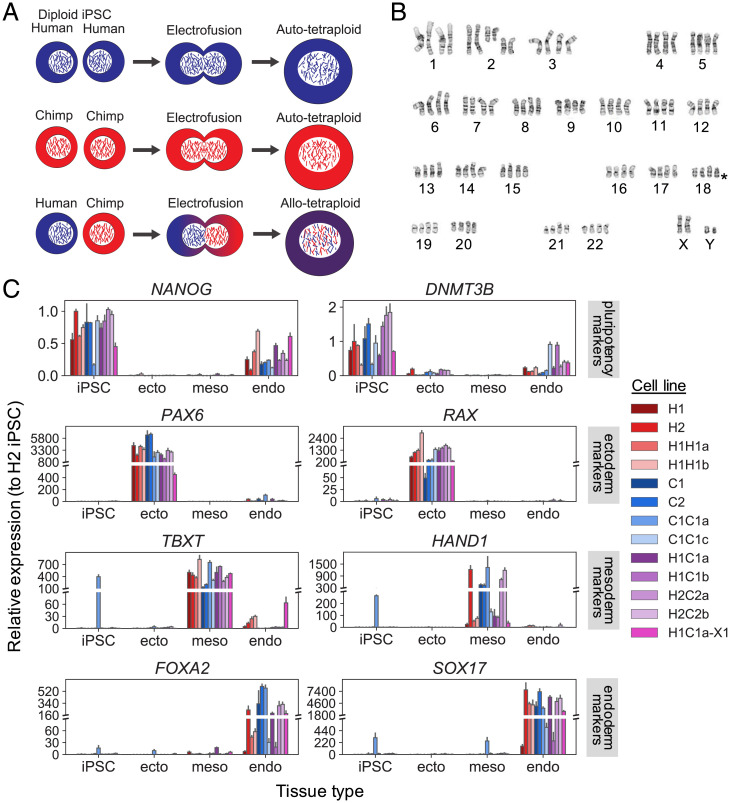
Fig. 1.
Generation and differentiation of autotetraploid and allotetraploid iPSC lines. Autotetraploid and allotetraploid cells contain the expected number of chromosomes and express expected marker genes after trilineage differentiation. (A) Human and chimpanzee diploid iPSC lines were labeled with diffusible dyes and subjected to electrofusion to generate autotetraploid and allotetraploid iPSC lines. (B) Tetraploid lines (H2C2a shown) exhibit karyotypes with four copies of each chromosome. Asterisk denotes location of a common iPSC human chr18q deletion (16), present in a subset of our cell lines. See Dataset S1 for detailed karyotype description of all lines. (C) Relative expression of pluripotency (NANOG, DNMT3B), ectoderm (PAX6, RAX), mesoderm (TBXT, HAND1), and endoderm (FOXA2, SOX17) marker genes tested via qRT-PCR after incubating cell lines under trilineage differentiation conditions. Cell lines tested are two human diploid lines (H1, H2), two human autotetraploid lines (H1H1a, H1H1b), two chimpanzee diploid lines (C1, C2), two chimpanzee autotetraploid lines (C1C1a, C1C1c), four allotetraploid lines (H1C1a, H1C1b, H2C2a, H2C2b), and one fluorescently marked allotetraploid line (H1C1a-X1). Gene expression is plotted relative to a human diploid undifferentiated iPSC line (H2). Error bars represent the SD of N = 3 cell culture replicates maintained as iPSCs or differentiated independently; 146 of 156 gene expression differences between undifferentiated cells and the tissue type in which a marker is expected to be expressed are significant by two-tailed Student’s t test at 5% FDR (see Dataset S3 for complete P value list).