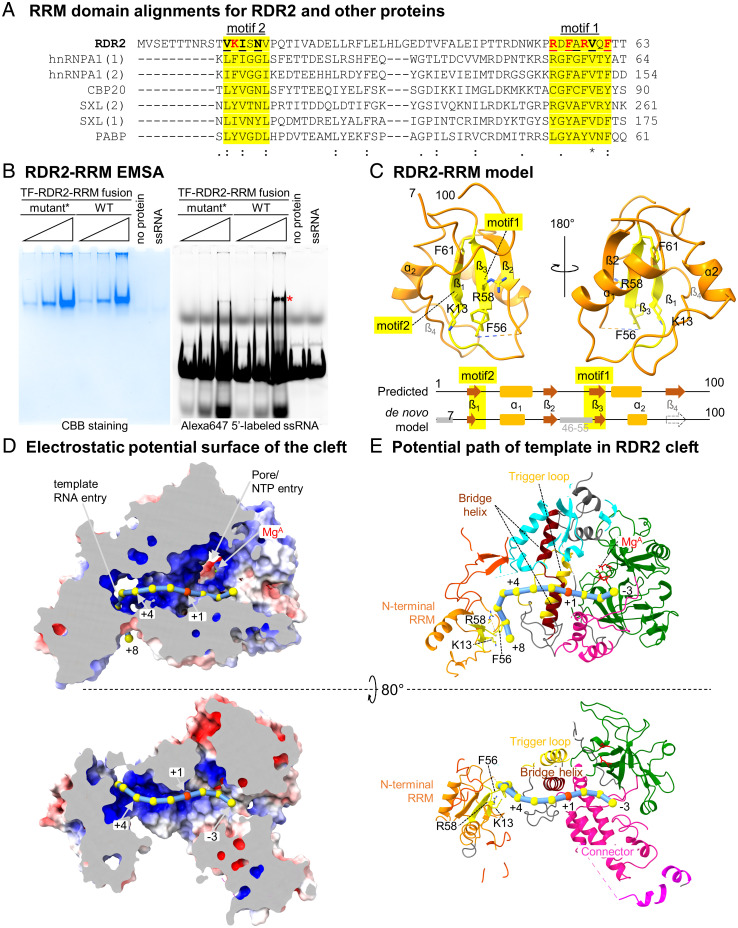
Fig. 3.
An N-terminal RRM domain influences the predicted path of template RNA. (A) RRM domain alignments for RDR2 (amino acids 1 to 63), human proteins: heterogeneous nuclear ribonucleoprotein A1 (hnRNPA), nuclear cap-binding protein subunit 2 (CBP) and polyA-binding protein 1 (PABP), and Drosophila melanogaster Sex lethal (SXL). Ribonucleoprotein 1 and 2 motifs (RNP1 and RNP2), having consensus sequences [RK]-G-[FY]-[GA]-[FY]-[ILV]-X-[FY] and [ILV]-[FY]-[ILV]-X-N-L, respectively, are shaded yellow. Basic or aromatic residues potentially interacting with RNA are shown in red. (B) EMSA of a recombinant TF-RDR2 (amino acids 1 to 100) fusion protein binding to a 37-nt RNA labeled with Alexa647. After acquiring the fluorescence image (Right), the gel was fixed and stained with Coomassie brilliant blue, CBB (Left). Wild-type and mutant forms of RDR2 were tested, with five amino acids of motifs 1 and 2 (shown in red in A) substituted by Ala in the mutant. An asterisk indicates the RNA–protein complex. (C) Structure of RDR2 (amino acids 1 to 100), with RRM domain motifs 1 and 2 colored yellow. The diagram at the bottom compares a computationally predicted secondary structure for the region to our structural model. (D) Electrostatic potential surface of RDR2 calculated using Adaptive Poisson–Boltzmann Solver (APBS) in Pymol with negative, neutral, and positive charges shown in red, white, and blue, respectively. RNA is modeled as beads on a string with the red bead indicating the position where complementary strand synthesis would begin (+1). Mg2+ and likely NTP entry pore positions are indicated. (E) Ribbon diagram views of the surface models shown in D. Domains are colored as in Fig. 1D. Additional views are shown in SI Appendix, Fig. S12.