Abstract
Perinatally HIV-infected children (PHIV), despite successful antiretroviral therapy, present suboptimal responses to vaccinations compared to healthy-controls (HC). Here we investigated phenotypic and transcriptional signatures of H1N1-specific B-cells (H1N1-Sp) in PHIV, differentially responding to trivalent-influenza-vaccine (TIV), and HC. Patients were categorized in responders (R) and non-responders (NR) according to hemagglutination-inhibition-assay at baseline and 21 days after TIV. No differences in H1N1-Sp frequencies were found between groups. H1N1-Sp transcriptional analysis revealed a distinct signature between PHIV and HC. NR presented higher PIK3C2B and NOD2 expression compared to R, confirmed by downregulation of PIK3C2B in resting-memory of R after H1N1 in-vitro stimulation. In conclusion this study confirms that qualitative rather than quantitative analyses are needed to characterize immune responses in PHIV. These results further suggest that higher PIK3C2B in H1N1-Sp of NR is associated with lower H1N1 immunogenicity and may be targeted by future modulating strategies to improve TIV responses in PHIV.
Keywords: Influenza vaccination, H1N1, Perinatally HIV infected children, PIK3C2B, Antigen specific B cells, Gene expression
1. Introduction
Despite effective antiretroviral therapy (ART), HIV infected patients present a suboptimal immunity to vaccines due to a depletion of central memory CD4+ T cells and memory B-cells [1–3]. We have previously reported lower serologic responses upon trivalent inactivated influenza vaccine (TIV) in perinatally HIV infected children (PHIV) [4,5]. We further defined transcriptional profiles of memory T [6] and B-cell subsets [7] associated to a lower TIV response in these patients. It is still unclear however, whether specific gene expression pathways of H1N1-specific B-cells (H1N1-Sp) rather than their frequency, may be responsible of reduced humoral immunity to H1N1 after TIV [8]. Overall, AKT, class I PI3K molecules and mTOR pathway seems to play a crucial role in the development of protective responses towards flu in humans [9]. This molecular signaling has been recently related to a direct suppression of entry and replication of Influenza A virus [10]. In addition, class I PI3K molecules were shown in mice to actively suppress class switch recombination and antibody-secreting cells in the early stage of the antibody response [11]. Following the hypothesis that this pathway within Ag specific B cells, could be involved in the Ab responses upon vaccination we recently reported from a single cell transcriptional profile approach, how PTEN, traditionally known as a class I PI3K inhibitor is upregulated in elderly HIV infected patients responding to TIV [12]. Recently PIK3C2B, a class II molecule, was shown to be involved in the AKT/mTOR signaling both in neuronal cells and in prostatic cancers [13–15]. To date no evidence have been reported linking the PI3K class II molecules to the Ag specific B cell function. With the final aim to dissect possible distinct transcriptional signatures of Ag specific B cells in samples collected from patients presenting differential responses upon TIV, we used a microfluidics Multiplex RT-PCR approach (Fluidigm, Biomark) for a curated gene panel (Supplementary Table 1), not limited only to the class II PI3K/AKT pathway. Here, we investigated the transcriptional profile of PI3K/AKT signaling pathway together with genes related to B cell home-ostasis, proliferation and differentiation on H1N1-specific B-cells (H1N1-Sp) of either PHIV under effective ART differentially responding to TIV or age-matched healthy controls (HC). Our results underline as PI3K/AKT signaling plays a pivotal role in persevering B cell dysfunction displayed by HIV-infected patients [16] leading to a suboptimal vaccine response.
2. Materials and methods
2.1. Study design
23 PHIV and 10 HC were enrolled at Bambino Gesù Children’s Hospital. Written informed consent was obtained from all subjects or parents/guardians of all minors for participation in a prospective open label influenza vaccine study. The study was conducted according to Good Clinical Practice guidelines and the Declaration of Helsinki. Bambino Gesù Children’s Hospital ethics committee approved the study. Only PHIV with a long-term viral control (≥ 24months) and good adherence to ART were considered eligible. Patients’ characteristics are described in Table 1. Participants were immunized between October 2012 and February 2013 with a single dose of seasonal 2012–2013 Inactivated Influenza Vaccine Trivalent Types A and B (Split Virion) VAXIGRIP® (Sanofi Pasteur). The strains included in the vaccine were A/California/7/2009 (H1N1) pdm09-like strain (abbreviated as H1N1), A/Victoria/361/2011 (H3N2)-like strain (abbreviated as H3N2), and B/Wisconsin/1/2010-like strain (abbreviated as B) as previously described [7]. PBMCs and plasma samples were collected at the time of immunization (T0) and 3 weeks after (T1), according to Food and Drug Administration Guidance for Industry (FDA, 2011) criteria. Vaccine responders (R) and non-responders (NR) were defined according to FDA (fda.gov).Patients with HAI titer to H1N1 at T1 of ≥1:40 and ≥ 4-fold increase compared to baseline were classified as R.Afterwards, additional samples (PBMCs) collected at T1 (from 19 PHIV and HC) were used to investigate H1N1-Sp in the present study.
Table 1.
Patients’ characteristics.
| HC | PHIV | R | NR | P value | |
|---|---|---|---|---|---|
| n | 8 | 19 | 9 | 10 | – |
| Gender (F) | 4 | 8 | 4 | 4 | n.s. |
| Age (years) (mean, SEM) | 18.8 (4.4) | 14.4 (1.3) | 14.9 (1.3) | 13.7 (2.5) | n.s. |
| HIV RNA < 50cp/mL (n) | – | 19 | 9 | 10 | – |
| H1N1 HAI fold change T1/T0 (mean, SEM) | 20 (1.15) | – | 19 (7.2) | 1 (0) | HC vs NR = ** R vs NR = ** |
| H1N1 HAI titer ≥ 1:40 at T0 | 6/8 | 17/19 | 8/9 | 9/10 | n.s. |
| Lymphocytes/μL (mean, SEM) | 2890 (194.72) | 2595 (222.6) | 2720 (173.6) | 3016 (359.1) | n.s. |
| CD4+ (mean, SEM) | 39.4 (3.9) | 44.5 (5.8) | 41.5 (8.7) | 50.3 (7.5) | n.s. |
| CD19+ (mean, SEM) | 7.9 (2.1) | 8.9 (1.6) | 7.9 (8.8) | 10.0 (2.9) | n.s. |
| CD19 + CD27 + IgD− (mean, SEM) | 13.3 (3.1) | 14.3 (2.1) | 12.7 (2.8) | 16.2 (3.3) | n.s. |
n, number of patients; HC, Healthy Controls; R, Responders; NR, Non Responders; F, Female; RM, Resting Memory; AM, Activated Memory; SEM, Standard Error of the Mean; n.s., p value>.5; ** = 0.0078).
2.2. H1N1 responses
Hemagglutination Inhibition Assay (HAI) test was performed and analyzed as previously described [7]. Briefly, plasma samples from all participants were pre-treated and diluted 1:4 in receptor-destroying enzyme (RDE) II (Boule Nordic AB) at 37 °C overnight, then incubated at 56 °C for 30 min. 4 H1N1 A hemagglutination units from H1N1 virus type A/California/7/2009 NYMC X-179A (GlaxoSmithKline) were incubated for 15 min at room temperature with an equal volume of RDE II treated plasma samples (in serial 2-fold dilutions, from 1:10 to 1:1280). After 50 μl of 0.5% erythrocytes from turkey were added to each sample. After 60 min the plate was acquired.
2.3. Flow cytometry, cells sorting and RNA extraction and multiplexed RT-PCR
PBMCs from T1 were thawed, resuspended in RPMI medium and stained with surface antibodies: CD3 (UCHT1), CD4 (SK3), CD8 (SK1), CD56 (NCAM16.2), CD10 (HI10a), CD14 (MφP9) (Dump channel, BV510, BD), CD19 ECD (HD237, Beckman Coulter), CD21 PeCy5 (Bly4, BD), CD27 BV605 (L128, BD), IgD PeCy7 (IA6-2, BD), gp140 (PE), H1N1 probe (PE), and Aqua marker to discriminate live/dead cells (BV510) and acquired using a four-way sorting mode on a FACS Aria II (BD). The gating strategy shown in Fig. 1A was used to determine B-cells subsets from total B, and to sort-purify a total of 100 cells per subset into different FACS tubes [6] containing CellsDirect One-Step PCR buffer and pooled TaqMan Gene Expression Assays (5 μl of 2× CellsDirect Reaction mix, 0.5 μl of Superscript III + Taq Polymerase, 2.5 μl of 0.2× TaqMan primer pool, and 1 μl of resuspension buffer). Samples were transferred into PCR tubes. H1N1 peptides, kindly provided by Sequirus (PA, USA) were used for in-vitro stimulation and reverse transcription, pre-amplification and multiplexed RT-PCR were performed as previously described [6]. Briefly pre-amplified samples were acquired on a 96.96 dynamic array IFC (Fluidigm). The pre-mix for the assay was made with 3 μl 20× TaqMan Gene Expression Assay (Applied Biosystems) and 3 μl 2× Assay Loading Reagent (Fluidigm). The pre-mix for the samples was made with 3 μl TaqMan Universal PCR Master Mix (2×, Applied Biosystems), 0.3 μl 20× GE Sample Loading Reagent (Fluidigm), and 2.7 μl of diluted cDNA. 5 μl of this preparation were loaded into the Fluidigm and results analyzed as in [6].
Fig. 1.
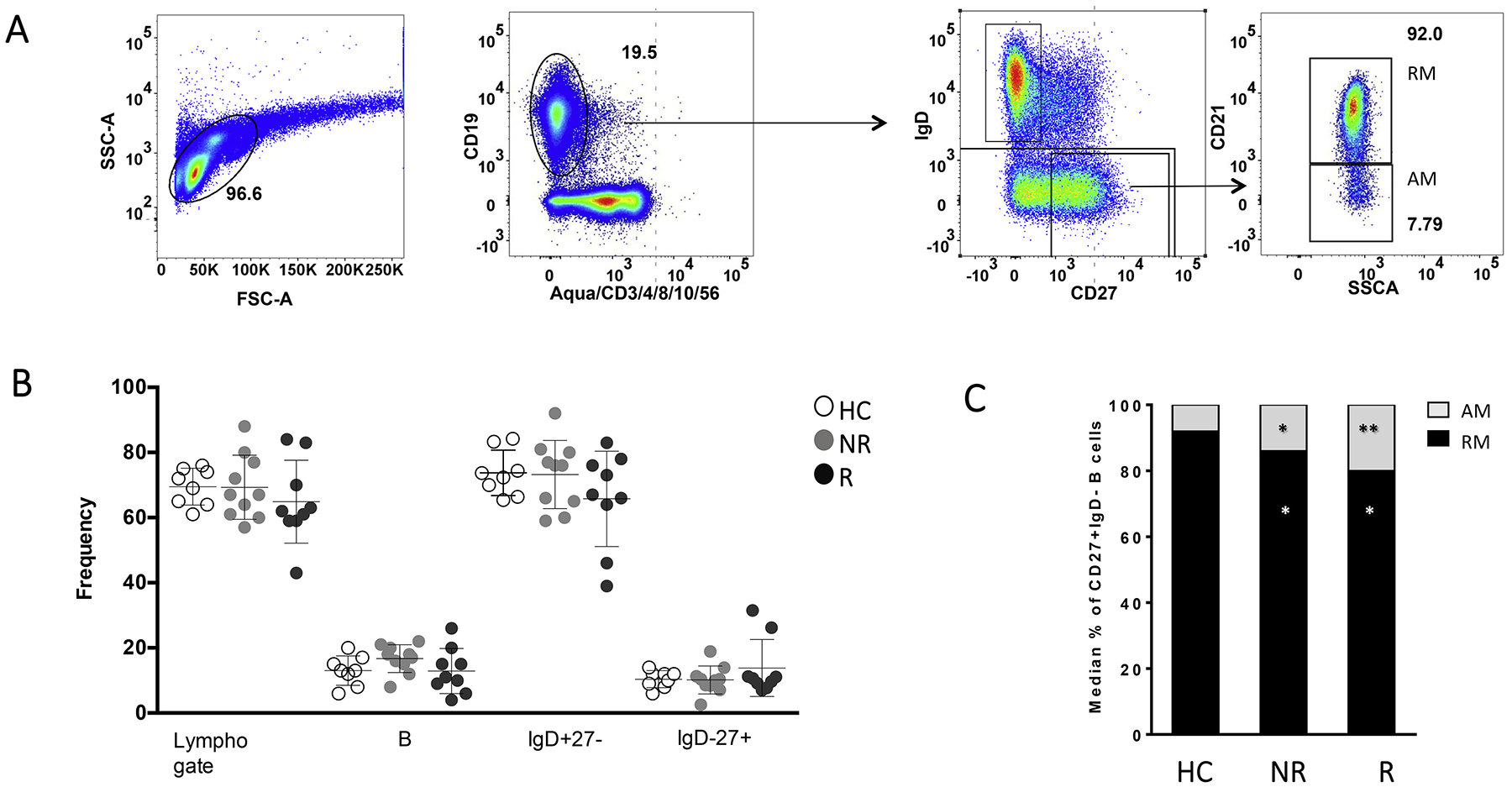
A) Gating strategy for general B cell subsets. B) Frequencies of total Lymphocytes, total B cells, IgD + CD27− (Naïve) and IgD-CD27+ (Switched memory) in healthy controls (HC), Non responder (NR) and Responder (R) patients. The Histogram in C) represent the median values of IgD-CD27+ cells, expressing (REM) or not expressing CD21 (AM). *, p values <0,04; **, p values <0,03 calculated between NR or R versus HC.
2.4. Statistical analysis
Outliers from gene expression analysis were filtered as follow: 1) removal of low Cellular Detection Rate depending on the dataset distribution; 2) removal of low expressed genes; and 3) removal of samples with extreme gene expression values (≥ +/− 7 standard deviation). Analysis were conducted through multi-contrast analysis pipeline using limma package for R (software R 3.0.2 GUI 1.62) as previously described [17] taking into account no H1N1 specific cells ANOVA analysis, performed by SingulaR (SingulaR analysis toolset 3.0), loaded on R allowed identification of differentially expressed genes (DEGs). Fold Change (FC) derived from mean Et values ere plotted on volcano plots, generated in Prism 6.0 (GraphPad). Only FC > 2.5 or < −2.5 with a p value <.05 was used to define DEGs. GSEA was performed with genemania.org as previously described [7]. Phosphatidylinositol signaling system pathway was created using the KEGG online tool KEGG Mapper – Search&Color Pathway.
3. Results
3.1. PHIV and HC present similar level of H1N1-specific B-cells despite their ability to respond to TIV
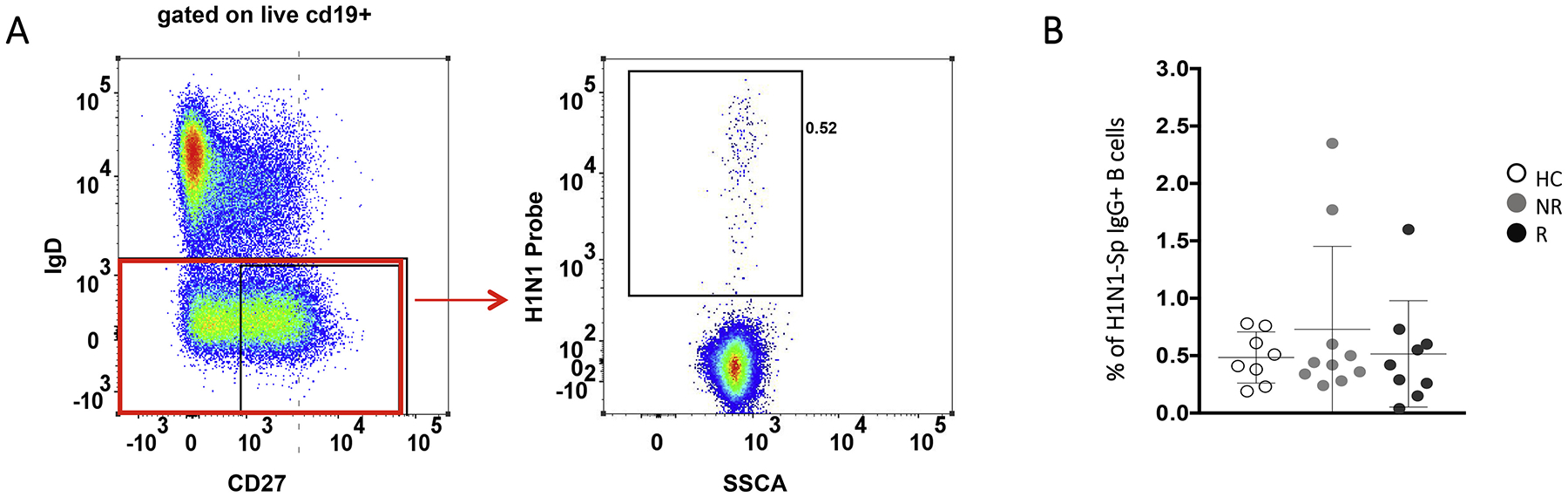
19 PHIV on effective ART and under viral suppression and 8 age-matched HC, previously enrolled in an influenza study [4] were randomly selected to investigate gene expression profiles and define possible biomarkers of TIV response. Responders (R) to TIV were individuals that showed at least 4-fold increase in H1N1 antibody titers (measured with Hemagglutination Inhibition Assay (HAI)) 21 days post-vaccination (T1) (Table 1) [4]. All HC fulfilled these criteria. No significant differences among the groups were found in terms of number of participants, gender, age, lymphocytes/μl, CD4+, CD19+, CD19 + CD27 + IgD−. All PHIV where under viral control (HIV RNA < 50 copies/mL) at the time of the study with a long history (>2 years) of viral control (Table 1). Flow cytometry analysis, performed on samples collected at T1, using a fluorescently labeled H1N1 probe and a panel of monoclonal antibodies, allowed us to identify H1N1-Sp (as previously described in [18]) (Fig. 1A–2A) and revealed no differences in terms of total B-cells, naive (IgD + CD27-) and switched memory (IgD-CD27+) B-cells (Fig. 1B). As we previously reported [7], HC showed higher frequency of CD21+ resting memory (RM: live, CD10−, CD19+ IgD− CD27+ CD21+) compared to R and non-responders (NR) (p = .035 and p = .04, respectively) while activated memory (AM: live, CD10−, CD19+ IgD− CD27+ CD21−) were significantly higher in PHIV for both R (p = .03) and NR (p = .023) when compared to HC (Fig. 1C). No differences were found between the groups in terms of frequency of H1N1-Sp among IgG+ switched memory B cells (Fig. 2B), frequency and absolute count of H1N1-Sp among total CD20+ B cells (data not shown) and B cell subsets distribution (data not shown).
Fig. 2.
A) Gating strategy used to identify B cell memory subsets and H1N1 specific cells. B) Dot plot depicting frequency of H1N1-Sp among IgG+ switched memory B cells in all groups.
3.2. PHIV H1N1-Sp present higher expression of genes related to apoptosis susceptibility and immune activation compared to HC
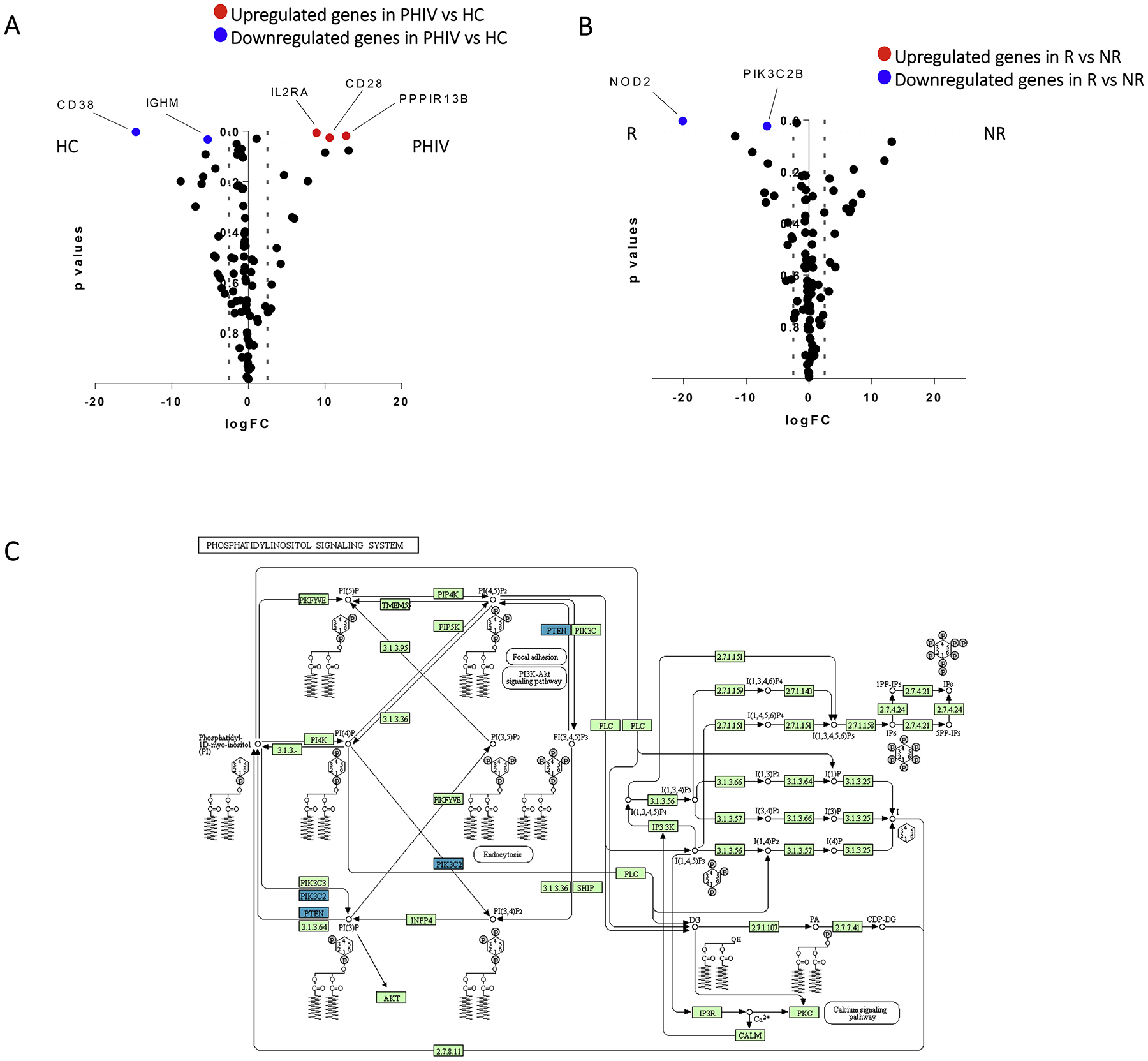
Equal numbers (100 cells, ~2 ng RNA) of live IgD-negative H1N1-sp were sort-purified and evaluated for transcript of a previously validated gene panel using the Fluidigm BioMark [7]. The multi-contrast analysis identified 5 DEGs comparing H1N1-sp of PHIV vs HC (Fig. 3A). Pathway analysis, applied to DEGs upregulated in PHIV (IL2RA, CD28 and PPPIR13B), pointed to immune activation and B-T lymphocyte interaction networks (data not shown). Indeed, IL2RA (logFC = 8.89, p = .005), a gene encoding for the subunit α of IL-2 receptor, and CD28 (logFC = 10.64, p = .02), play a central role in the B-T cells interactions towards activation and proliferation. Also, the upregulation of PPP1R13B (logFC = 12.77, p = .01), which encodes for a member of the ASPP (apoptosis-stimulating protein of p53) family of p53 interacting proteins, suggests a gene profile prone to exhaustion and activation in H1N1-sp of PHIV compared to HC. In line with this, other molecules suggested by the GSEA (not shown) and actively involved in the regulation of such DEGs were CD80, CD86, IL2, CTLA4, ICAM1. On the other hand, the down-regulated DEGs in PHIV were CD38 (logFC = −14.680, p = .002) and IGHM (logFC = −5.30, p = .03) both coding for protein involved in proliferation and IgM within mechanisms of primary and low specificity defense responses upon infection.
Fig. 3.
A) Volcano plot representing Differentially Expressed Genes (DEGs) with a significant p value, comparing the expression in PHIV versus HC; red dots show upregulated genes in PHIV, while blue dots downregulated ones. B) Volcano plot depicting in blue downregulated DEGs with a significant p value in R vs NR patients; no upregulated genes were found. C) Phosphatidylinositol signaling system pathway depicting in blue downregulated DEGs with a significant p value in R vs NR patients.
3.3. PIK3C2B is downregulated in H1N1-Sp of R compared to NR, and in RM of R, after H1N1 in-vitro stimulation
Multi contrast analysis of H1N1-Sp was performed in PHIV differentially responding to TIV in order to define specific profiles associated to H1N1 response after TIV. A 2 DEGs signature showing down-regulation of NOD2 (logFC = −20.11, p = .003) and PIK3C2B (logFC = −6.67, p = .02) was found in R compared to NR (Fig. 3B).Also in Fig. 3 C the phosphatidylinositol signaling system pathway is reported, showing in blue the down-regulated DEGs in R versus NR. Gene Set Enrichment Analysis (GSEA) revealed that top related networks to these DEGs are mainly involved in the activation during inflammation or activation mechanisms following surface antigen (Ag) recognition (data not shown) and directed towards NFkB activation. Other genes associated to the pathway were PI3K subunits (PIK3CB, PIK3R1, PIK3R2, PIK3R3) and additional activation markers such as CD28 and IKBKG that regulate NFkB activity.
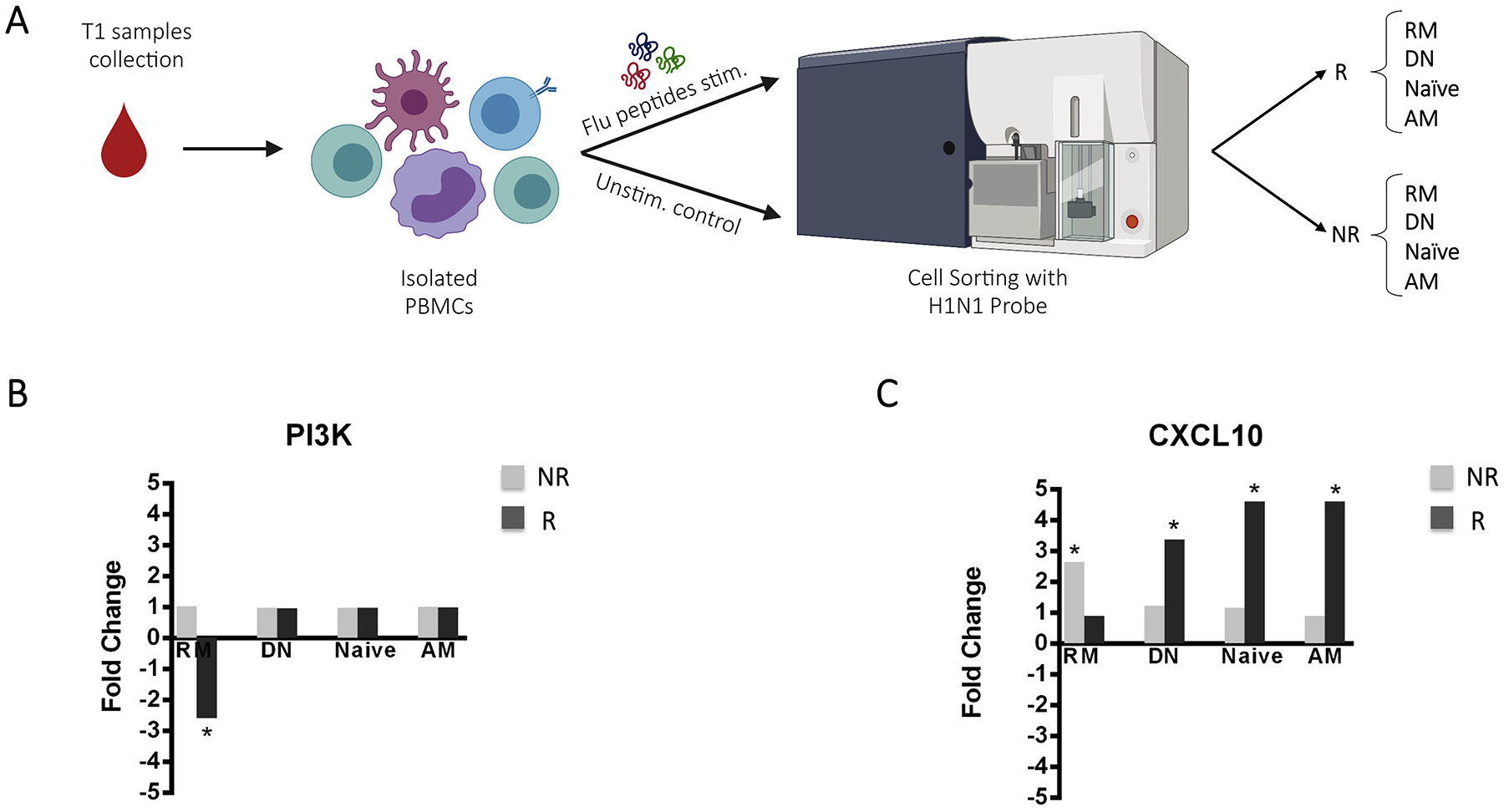
To further investigate the role of PIK3C2B in R and NR, an additional aliquot of PBMCs collected from these patients at T1, was stimulated in-vitro with H1N1 peptides and multiplexed RT-PCR was performed on B-cell subsets as previously described [6] (Fig. 4A). Interestingly, PIK3C2B showed down regulation in in-vitro stimulated samples of R when compared to unstimulated in the RM subset (Fig. 4B). No differences were found in NR in terms of PIK3C2B gene expression variation after in-vitro stimulation in NR B-cell subsets. Interestingly, CXCL10 a downstream molecule of the PIK3C2B pathway was upregulated in RM only of NR. On the other hand, R showed upregulation of CXCL10 in AM, double negative (DN: live, CD10−, CD19+, IgD−, CD27−) and Naïve B-cells (Naïve: live, CD10−, CD19+, IgD+, CD27−), presumably suggesting that this activation in non-RM B-cells is PIK3C2B-independent after in-vitro stimulation (Fig. 4C).
Fig. 4.
A) Cartoon depicting the strategy used to sort-purify 100 cells in 4 different subpopulations for both R and NR (Created with Biorender.com). B-C) The two histograms represent the fold change expression of B) PIK3C2B and C) CXCL10, respectively among B cell subpopulations (RM, DN, Naive and AM) (* represents p values lower than 0.05) after in-vitro stimulation.
4. Discussion
PHIV present a suboptimal serological and cellular response upon immunization [19]. With regards to Influenza vaccination, we have previously reported distinct B-cell signatures in PHIV differentially responding to TIV [4,7]. It is still unclear, however, whether such differences could be driven by qualitative rather than quantitative disturbances of memory B-cells involved in the development of protective immune response towards H1N1 virus. In the present study, we show that no differences exist in terms of frequency, absolute counts and B-cell subsets distribution of H1N1-Sp between PHIV and HC. In addition, no differences were found between R and NR. These data are in line with ART-treated HIV infected adults after TIV [20,21]. AM were found significantly higher in PHIV (both R and NR) compared with HC. AM (CD19+ IgD− CD27+ CD21−) have been described in the HIV scenario to be associated to a chronic exposure to HIV viremia in chronically HIV-infected patients and to dominate abnormal HIV specific responses [22]. In the past few years we and other groups have shown that despite long term viral control perturbation at both frequency and transcriptional level persist in vertically HIV infected children in activated memory B cells [7]. This subset is also expanded in other models of primary immune deficiencies characterized by recurrent infection and chronic immune activation such as chronic granulomatous disease [23].Also mature activated B cells (CD10-CD21-), analyzed regardless the expression of the memory molecule CD27, were shown to be correlated to a lower H1N1 response upon trivalent inactivated vaccination [4].
To further dissect whether specific gene programs, related to antibody-secreting cells (ASCs) proliferation, and class switch recombination (CSR) may determine a lower serologic response in NR we characterized at a transcriptional level purified H1N1-Sp from both PHIV (R and NR) and HC. For our transcriptional analysis we used the Fluidigm Biomark platform, in order to interrogate gene expression of a limited and curated panel of 96 genes from Ag specific and non-Ag specific B-cell subsets. Although this approach represents a limit compared with the unbiased whole-transcriptome analyses (e.g., RNA sequencing or microarray), it allowed to obtain a large amount of transcriptional information on highly relevant cell types such as Ag specific B- cells and from a relatively low cell numbers obtained from pediatric samples [24].
In line with our previous reports [7,24], comparisons between H1N1-Sp of HC and PHIV, revealed a higher expression of genes involved in immune activation and inflammation (CD28, IL2RA, and PPP1R13B) in PHIV. These data suggest that transcriptional perturbation persists at Ag specific level in these patients despite long term effective ART and viral control.
In the present study we further aimed at dissecting transcriptional pathways of H1N1-Sp involved in immune responses upon Ag stimulation possibly informing the ability of PHIV to respond to TIV. Whereas specific genes such as BLIMP1, BCL6 or AID have been shown to drive the B cell towards ASCs, CSR or Germinal Center reactions, fewer evidence has been reported on signaling pathways involved in such mechanisms, especially at the peripheral blood level and in Ag-specific B-cells [25]. Different models have proposed that class I PI3K activation is intimately linked to ASC and CSR in B-cells [26]. Indeed, it prevents premature CSR during the early phases of the Ag encountering [11] thus reducing over clonal expansion when high Ag loads link to the B-cell receptor. Fewer studies have been proposed with respect to the class II molecules of PI3K. Interestingly these molecules, such as PIK3C2B, have been shown to be involved in the AKT/mTOR signaling pathway in other cell lines [13,27,28] and no evidence, to date has been produced linking such pathway with B cell function. In the present study, multi-contrast analysis from gene expression data deriving from H1N1-Sp comparisons from samples collected 21 days post TIV revealed a higher expression of PIK3C2B in NR compared to R, suggesting a lower ability of such cells to induce ASCs in a PIK3C2B-inhibition-dependent manner. We found, for the first time in the context of pediatric HIV infection, a distinct signature in this pathway exclusively within RM subset, where the highest affinity memory responses reside. Indeed, PIK3C2B was downregulated only in samples of R, as observed after TIV and confirmed by in-vitro stimulation with H1N1 peptides. In addition,CXCL10, showed to be triggered by the PI3K activation [29], resulted positively induced after in-vitro H1N1 stimulation in RM of NR further suggesting the lack of inhibitory role of PIK3C2B in this specific memory B cell subset in PHIV NR. This is in line with recent findings presented in a cohort of horizontally HIV-infected elderly where PTEN, a PIK3 inhibitor, was found upregulated in H1N1 responders [12]. The importance of this pathway is further supported by a mice study showing that the Ag specific ASC induced by Blimp1 after Ag stimulation is partially dependent on PI3K modulation [11]. Another DEG, found upregulated in NR vs R was NOD2. NOD2 codes for a protein that is mainly expressed in pheripheral blood leukocytes and can induce the activation of the NF-κB or MAPK (through the interaction with Receptor-interactin protein 2 (RIP2)) pathways to induce inflammatory reaction [30–32]. It has also been recently described to be a negative regulator of the TLR2/TLR4 driven response [32]. The activation of both TLR2 and TLR4 has been strongly linked to proliferation and ASCs production after in vivo and in-vitro stimulation [30]. These results, may suggest that the NOD2, found upregulated in NR, leads to an inhibitory effect on the TLR2/TLR4 B cell driven response in H1N1-Sp after TIV thus hampering the capability of these cells to undergo effective ASCs production and proliferation. Overall, these results may suggest that the ability to control inflammation signals trough PIK3C2B and NOD 2 downregulation within H1N1-Sp resting memory B-cells is needed in order to provide a serologic and cellular response upon TIV in PHIV (depicted in graphical abstract). A gain of function mutation in class I phosphoinositide 3-kinase δ (PI3Kδ) was recently associated to the Activated PI3-kinase delta syndrome (APDS), characterized by defective antibody responses, inflammation and autoimmunity [33]. Since this field still remain poorly investigated, deep studies on class II PI3K molecules such as PIK3C2B are needed in order to understand if a similar mechanism as with PI3Kδ could be happening in NR PHIV.
In conclusion, the present study suggests that a 2 DEGs signature of H1N1-Sp B-cells of PHIV is associated to TIV immunogenicity. Upregulation of PIK3C2B, found in PHIV NR, may suggest the future use of vaccine-adjuvants aimed at specifically modulating PIK3C2B activity, especially considering that specific compounds are currently under investigation for the APDs [34]. This study suggests that specific transcriptional signatures of Ag-specific B-cells rather than their frequency, provide crucial information on the ability of PHIV to respond to Influenza vaccination and should be further investigated by larger studies.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.clim.2020.108440.
Supplementary Material
Acknowledgements
We would like to acknowledge all patients and guardians who decided to participate to the study. We thank Celeste Sanchez for her help in experimental work and in phase of analysis. We acknowledge Giuseppe Pascucci for his precious help with gene ontology analysis. We thank Rajendra Pahwa for helpful suggestions and Jennifer Faudella for her administrative assistance. We thank Nadia Iavarone and Tamara Di Marco for clinical assistance.
Funding
This work was made possible by the support from a pilot award to NC from Miami Center for AIDS Research (CFAR), grants obtained by Bambino Gesú Children’s Hospital (Ricerca corrente 2020 to NC), Associazione Volontari Bambino Gesù, Ricerca Finalizzata 2010, Ministero della Salute (RF_2010_2310438), and grants AI108472 and AI127347 to SP and the Laboratory Sciences Core of the Miami CFAR (P30AI073961) from the National Institutes of Health (NIH), which is supported by the following NIH Co-Funding and Participating Institutes and Centers: NIAID, NCI, NICHD, NHLBI, NIDA, NIMH, NIA, NIDDK, NIGMS, FIC, and OAR.
Footnotes
Financial disclosure
The authors have no financial relationship to this article to disclose.
Declaration of Competing Interest
The authors have declared that no conflict of interest exists.
References
- [1].Pallikkuth S, Parmigiani A, Silva SY, George VK, Fischl M, Pahwa R, Pahwa S, Impaired peripheral blood T-follicular helper cell function in HIV-infected non-responders to the 2009 H1N1/09 vaccine, Blood 120 (2012) 985–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Titanji K, De Milito A, Cagigi A, Thorstensson R, Grutzmeier S, Atlas A,Hejdeman B, Kroon FP, Lopalco L, Nilsson A, Chiodi F, Loss of memory B cells impairs maintenance of long-term serologic memory during HIV-1 infection, Blood 108 (2006) 1580–1587. [DOI] [PubMed] [Google Scholar]
- [3].Paolo P, Rinaldi S, Cotugno N, Santilli V, Pahwa S, Rossi P, Cagigi A, Premature B-cell senescence as a consequence of chronic immune activation Implications for vaccination of immune compromised individuals, Human Vaccines & Immunotherapeutics 10 (2014) 2083–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cagigi A, Rinaldi S, Di Martino A, Manno EC, Zangari P, Aquilani A,Cotugno N, Nicolosi L, Villani A, Bernardi S, Donatelli I, Pahwa S, Rossi P,Palma P, Premature immune senescence during HIV-1 vertical infection relates with response to influenza vaccination, J. Allergy Clin. Immunol 133 (2014) 592–594. [DOI] [PubMed] [Google Scholar]
- [5].Stefano R, Rinaldi S, Zangari P, Cotugno N, Manno EC, Brolatti N,Castrucci MR, Donatelli I, Rossi P, Palma P, Cagigi A, Antibody but not memory B-cell responses are tuned-down in vertically HIV-1 infected children and young individuals being vaccinated yearly against influenza, Vaccine 32 (2014) 657–663. [DOI] [PubMed] [Google Scholar]
- [6].De Armas LR, Cotugno N, Pallikkuth S, Pan L, Rinaldi S, Sanchez MC,Gonzalez L, Cagigi A, Rossi P, Palma P, Pahwa S, Induction of IL21 in peripheral T follicular helper cells is an indicator of influenza vaccine response in a previously vaccinated HIV-infected pediatric cohort, J. Immunol 198 (2017) 1995–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cotugno N, De Armas L, Pallikkuth S, Rinaldi S, Issac B, Cagigi A, Rossi P,Palma P, Pahwa S, Perturbation of B cell gene expression persists in HIV-infected children despite effective antiretroviral therapy and predicts H1N1 response, Front. Immunol 8 (2017) 1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cotugno N, Ruggiero A, Santilli V, Manno EC, Rocca S, Zicari S, Amodio D,Colucci M, Rossi P, Levy O, Martinon-Torres F, Pollard AJ, Palma P, OMIC Technologies and Vaccine Development: From the Identification of Vulnerable Individuals to the Formulation of Invulnerable Vaccines, J Immunol Res 8732191 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Garcia CC, Tavares LP, Dias ACF, Kehdy F, Alvarado-Arnez LE, Queiroz-Junior CM, Galvao I, Lima BH, Matos AR, Goncalves APF, Soriani FM,Moraes MO, Marques JT, Siqueira MM, Machado AMV, Sousa LP, Russo RC, Teixeira MM, Phosphatidyl inositol 3 kinase-gamma balances antiviral and inflammatory responses during Influenza A H1N1 infection: from murine model to genetic association in patients, Front. Immunol 9 (2018) 975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hirata N, Suizu F, Matsuda-Lennikov M, Edamura T, Bala J, Noguchi M, Inhibition of Akt kinase activity suppresses entry and replication of influenza virus, Biochem. Biophys. Res. Commun 450 (2014) 891–898. [DOI] [PubMed] [Google Scholar]
- [11].Omori SA, Cato MH, Anzelon-Mills A, Puri KD, Shapiro-Shelef M, Calame K, Rickert RC, Regulation of class-switch recombination and plasma cell differentiation by phosphatidylinositol 3-kinase signaling, Immunity 25 (2006) 545–557. [DOI] [PubMed] [Google Scholar]
- [12].De Armas LR, Pallikkuth S, Pan L, Rinaldi S, Cotugno N, Andrews S, Pahwa R, Mcdermott AB, Palma P, Pahwa S, Single cell profiling reveals PTEN over-expression in influenza-specific B cells in aging HIV-infected individuals on antiretroviral therapy, Sci. Rep 9 (2019) 2482 Fda (2011). Available: http://www.fda.gov/media/73339/download [Accessed]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Das M, Scappini E, Martin NP, Wong KA, Dunn S, Chen YJ, Miller SL, Domin J, O’bryan JP, Regulation of neuron survival through an intersectinphosphoinositide 3′-kinase C2beta-AKT pathway, Mol. Cell. Biol 27 (2007) 7906–7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gulluni F, De Santis MC, Margaria JP, Martini M, Hirsch E, Class II PI3K functions in cell biology and disease, Trends Cell Biol 29 (2019) 339–359. [DOI] [PubMed] [Google Scholar]
- [15].Mavrommati I, Cisse O, Falasca M, Maffucci T, Novel roles for class II Phosphoinositide 3-kinase C2beta in signalling pathways involved in prostate cancer cell invasion, Sci. Rep 6 (2016) 23277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Moir S, Fauci AS, Pathogenic mechanisms of B-lymphocyte dysfunction in HIV disease, J. Allergy Clin. Immunol 122 (2008) 12–19 (quiz 20–11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK, Limma powers differential expression analyses for RNA-sequencing and microarray studies, Nucleic Acids Res 43 (2015) e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Whittle JR, Wheatley AK, Wu L, Lingwood D, Kanekiyo M, Ma SS,Narpala SR, Yassine HM, Frank GM, Yewdell JW, Ledgerwood JE, Wei CJ, Mcdermott AB, Graham BS, Koup RA, Nabel GJ, Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages, J. Virol 88 (2014) 4047–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Obaro SK, Pugatch D, Luzuriaga K, Immunogenicity and efficacy of childhood vaccines in HIV-1-infected children, Lancet Infect. Dis 4 (2004) 510–518. [DOI] [PubMed] [Google Scholar]
- [20].Sun P, Crum-Cianflone NF, Defang G, Williams M, Ganesan A, Agan BK,Lalani T, Whitman T, Brandt C, Burgess TH, Evaluation of T and B memory cell responses elicited by the pandemic H1N1 vaccine in HIV-infected and HIV-uninfected individuals, Vaccine 35 (2017) 6103–6111. [DOI] [PubMed] [Google Scholar]
- [21].Wheatley AK, Kristensen AB, Lay WN, Kent SJ, HIV-dependent depletion of influenza-specific memory B cells impacts B cell responsiveness to seasonal influenza immunisation, Sci. Rep 6 (2016) 26478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kardava L, Moir S, Shah N, Wang W, Wilson R, Buckner CM, Santich BH,Kim LJ, Spurlin EE, Nelson AK, Wheatley AK, Harvey CJ, Mcdermott AB, Wucherpfennig KW, Chun TW, Tsang JS, Li Y, Fauci AS, Abnormal B cell memory subsets dominate HIV-specific responses in infected individuals, J. Clin. Invest 124 (2014) 3252–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cotugno N, Finocchi A, Cagigi A, Di Matteo G, Chiriaco M, Di Cesare S, Rossi P,Aiuti A, Palma P, Douagi I, Defective B-cell proliferation and maintenance of long-term memory in patients with chronic granulomatous disease, J. Allergy Clin. Immunol 135 (753–761) (2015) e752. [DOI] [PubMed] [Google Scholar]
- [24].Cotugno N, De Armas L, Pallikkuth S, Rossi P, Palma P, Pahwa S, Paediatric HIV infection in the ‘omics era: defining transcriptional signatures of viral control and vaccine responses, J. Virus Erad 1 (2015) 153–158. [PMC free article] [PubMed] [Google Scholar]
- [25].Cotugno N, Morrocchi E, Rinaldi S, Rocca S, Pepponi I, di Cesare S, Bernardi S,Zangari P, Pallikkuth S, de Armas L, Levy O, Rossi P, Pahwa S, Palma P, EPIICAL Group, Early antiretroviral therapy-treated perinatally HIV-infected seronegative children demonstrate distinct long-term persistence of HIV-specific T-cell and B-cell memory, AIDS 34 (5) (2020) 669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Werner M, Hobeika E, Jumaa H, Role of PI3K in the generation and survival of B cells, Immunol. Rev 237 (2010) 55–71. [DOI] [PubMed] [Google Scholar]
- [27].Alliouachene S, Bilanges B, Chicanne G, Anderson KE, Pearce W, Ali K, Valet C,Posor Y, Low PC, Chaussade C, Scudamore CL, Salamon RS, Backer JM,Stephens L, Hawkins PT, Payrastre B, Vanhaesebroeck B, Inactivation of the class II PI3K-C2beta potentiates insulin signaling and sensitivity, Cell Rep 13 (2015) 1881–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Margaria JP, Ratto E, Gozzelino L, Li H, Hirsch E, Class II PI3Ks at the intersection between signal transduction and membrane trafficking, Biomolecules 9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kaur S, Sassano A, Joseph AM, Majchrzak-Kita B, Eklund EA, Verma A,Brachmann SM, Fish EN, Platanias LC, Dual regulatory roles of phosphatidylinositol 3-kinase in IFN signaling, J. Immunol 181 (2008) 7316–7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bekeredjian-Ding I, Jego G, Toll-like receptors–sentries in the B-cell response, Immunology 128 (2009) 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Boyle JP, Parkhouse R, Monie TP, Insights into the molecular basis of the NOD2 signalling pathway, Open Biol 4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kim H, Zhao Q, Zheng H, Li X, Zhang T, Ma X, A novel crosstalk between TLR4-and NOD2-mediated signaling in the regulation of intestinal inflammation, Sci. Rep 5 (2015) 12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Angulo I, Vadas O, Garcon F, Banham-Hall E, Plagnol V, Leahy TR, Baxendale H, Coulter T, Curtis J, Wu C, Blake-Palmer K, Perisic O, Smyth D, Maes M, Fiddler C, Juss J, Cilliers D, Markelj G, Chandra A, Farmer G, Kielkowska A, Clark J, Kracker S, Debre M, Picard C, Pellier I, Jabado N, Morris JA, Barcenas-Morales G, Fischer A, Stephens L, Hawkins P, Barrett JC, Abinun M, Clatworthy M, Durandy A, Doffinger R, Chilvers ER, Cant AJ, Kumararatne D, Okkenhaug K, Williams RL, Condliffe A, Nejentsev S, Phosphoinositide 3-kinase delta gene mutation predisposes to respiratory infection and airway damage, Science 342 (2013) 866–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Coulter TI, Cant AJ, The treatment of activated PI3Kdelta syndrome, Front. Immunol 9 (2018) 2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


