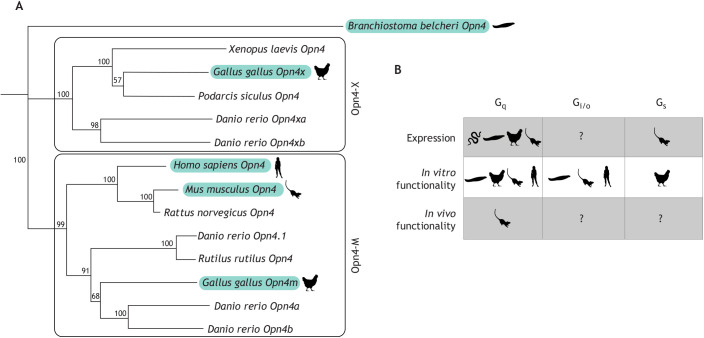
Fig. 4.
Phylogeny and current understanding of melanopsin phototransduction across species. (A) Phylogenetic analysis of the melanopsin protein sequence across multiple organisms using the neighbor-joining method (Saitou and Nei, 1987). Amino acid sequences of 14 melanopsin genes expressed in 9 different organisms (UniProt, Table S1) were aligned using MUSCLE alignment (Edgar, 2004) and compiled into a tree using the neighbor-joining method within Geneious Prime (Geneious Prime v.2019.1.1.; https://www.geneious.com). Amphioxus (Branchiostoma belcheri) melanopsin is used as an outgroup. Branches are labeled with percent confidence levels based on 100 bootstrap replicates. Only the melanopsin(s) of highlighted species have been studied in the context of phototransduction. (B) A summary of the current knowledge of the cognate G-proteins of melanopsin across multiple organisms. ‘Expression’ denotes either expression of protein or expression of mRNA transcripts that co-localize with melanopsin. Expression and in vitro assessment of G-protein activation has been defined across multiple organisms, whereas in vivo work has been done exclusively in the mouse.