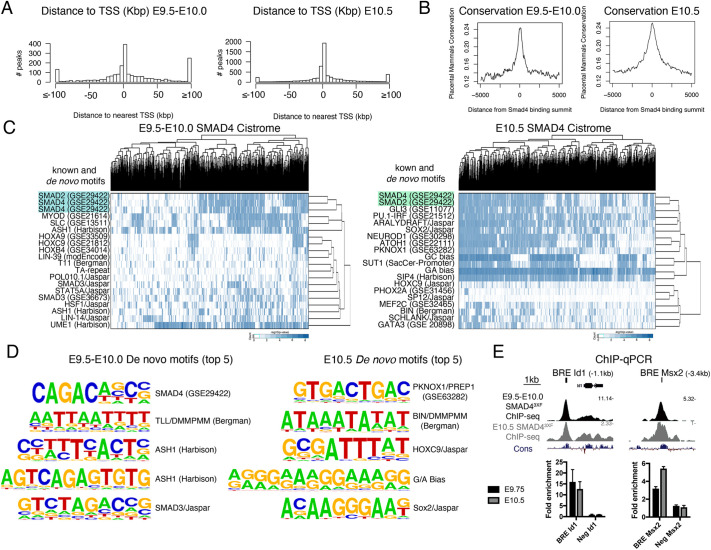
Fig. 1.
Identification of the genomic regions enriched in SMAD4-chromatin complexes in mouse forelimb buds. (A) Histogram showing the distribution of SMAD4-interacting regions in relation to the nearest transcriptional start site (TSS) at E9.5-E10.0 (25-30 somites) and E10.5 (34-38 somites). (B) The average Phastcons conservation of the genomic regions enriched in SMAD4-chromatin complexes is shown at both stages. (C) Hierarchical clustering of the high-affinity matches for the top known and de novo motifs enriched in the SMAD4-bound regions at both stages. (D) The top five de novo motif identified in the genomic regions enriched in SMAD4-chromatin complexes in forelimb buds at both stages. (E) ChIP-qPCR validation of two previously known SMAD4-interacting genomic regions – BREs for Id1 and Msx2, respectively – at both stages. Two biological replicates were analyzed (data are mean±s.d. of three technical replicates).