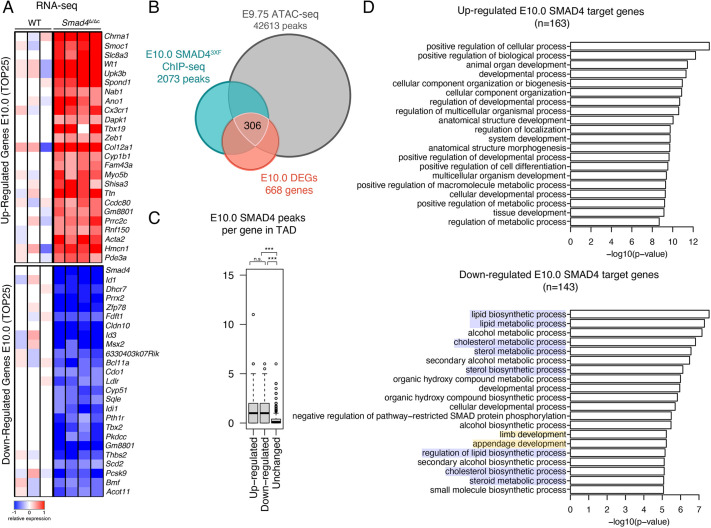
Fig. 2.
Identification of SMAD4 target genes in mouse forelimb buds at E10.0. (A) Heatmaps showing the DEGs identified by comparing wild-type (WT) and Smad4Δ/Δc transcriptomes at E10.0 (30 somites; wild type, n=3; Smad4Δ/Δc, n=4 biological replicates). The top 25 up- and downregulated genes are shown. For each gene, the log2-ratio between the expression level in each sample and the mean of the three biological replicates for the wild-type forelimb buds is shown. Red indicates increased expression and blue indicates reduced expression in comparison with the mean of the wild-type samples. (B) Three-way Venn diagram showing the intersection between the ChIP-seq (E9.5-10.5, 25-30 somites), ATAC-seq (E9.75, 26 somites) and RNA-seq (E10.0, 30 somites) datasets. This identifies 306 candidate SMAD4 target genes in mouse forelimb buds. (C) Box plot representing the number of E9.5-E10.0 SMAD43xF ChIP-seq peaks within a TAD of genes that are either up- or downregulated in Smad4Δ/Δc forelimb buds in comparison with genes with unaltered expression. The box plot indicates median (50th percentile) and interquartile range (25th and 75th percentiles; IQR) with whiskers set at 1.5×IQR. Values exceeding the whiskers are considered outliers and are individually marked. Upregulated versus unchanged, P=5.2e-26; downregulated versus unchanged, P=9.9e-32 (Mann–Whitney test). n.s., not significant. (D) GO enrichment analysis of biological processes of the up- and downregulated SMAD4 target genes in Smad4Δ/Δc forelimb buds. The GO terms for processes relevant to sterol/lipid/sterol/cholesterol biosynthesis and to limb development are highlighted in blue and yellow, respectively (bottom panel).