Figure 1.
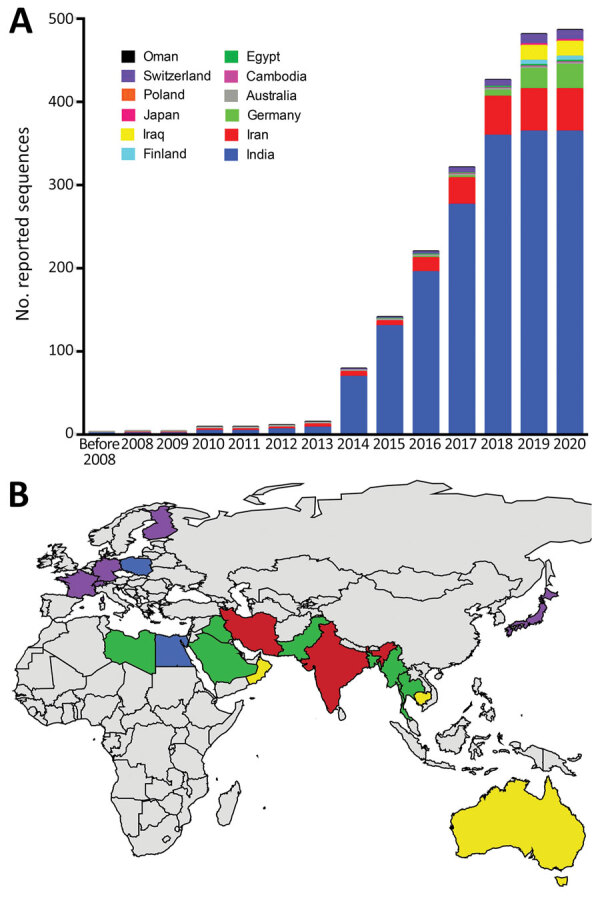
Analysis of dynamic and geographic distribution of Trichophyton indotineae reported sequences from France (this study) and reference sequences from GenBank for 2004–2021 A) Cumulative curves of 486 published sequences; B) geographic distribution of 537 published sequences. Red, countries with reported endemic cases; purple, countries with imported cases (but rare cases of endemic transmission cannot be ruled out); green, probable country sources of imported cases; yellow, countries with reported sporadic human cases without additional available information (also identified in Poland); blue, countries with T. indotineae sequences reported in animal infections (also reported in India). World map was created using JMP Pro 15.2.0 (https://www.jmp.com). For internal transcribed spacer sequence-based screening, we retrieved ITS1-5.8S-ITS2 sequences T. interdigitale, T. mentagrophytes, T. indotineae and also Anthroderma benhamiae, A. simii, A. vanbreuseghemii, T. benhamiae, T. bullosum, T. concentricum, T. equinum, T. erinacei, T. quinckeanum, T. simii, T. schoenleinii, T. tonsurans, and T. verrucosum. For sequences matching T. indotineae (internal transcribed spacer reference sequence JN133999), we searched associated literature on PubMed Central (https://www.ncbi.nlm.nih.gov/pmc).
