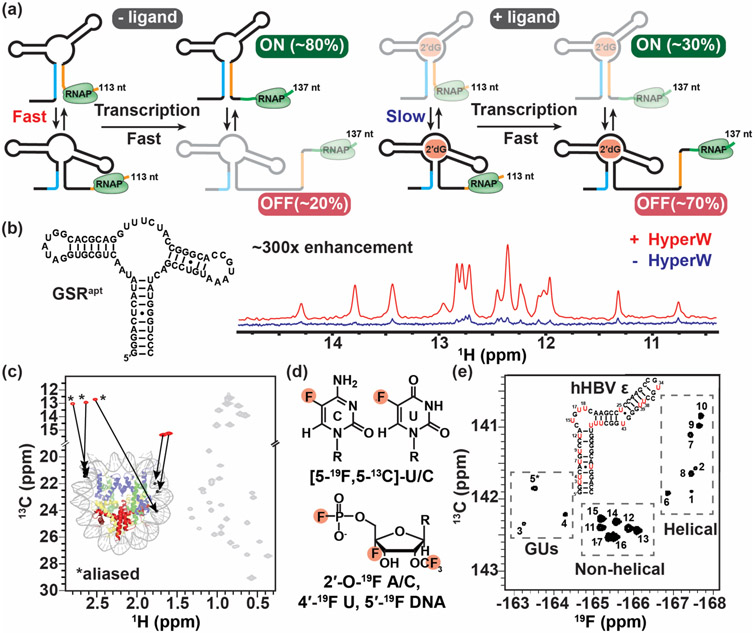
Figure 4.
Real-time NMR and methods to study large nucleic acid assemblies. (a) Schematic representation of the interconverting transcriptional intermediates of 2′dG-sensing riboswitch at transcript lengths 113 and 137 nt with and without ligand. Ligand binding slows the conformational exchange between the OFF and ON state below the transcription rate, triggering the OFF state function. The figure was adapted from [60]. (b) 1D imino spectra of the guanine-sensing riboswitch (GSR) aptamer domain (GSRapt) of the Bacillus subtilis xpt-pbuX operon with (red) and without (blue) injection of hyperpolarized water (HyperW). The figure was adapted from [62]. (c) The 2D 13C-1H HMQC spectrum of a nucleosome core particle (NCP) (shown is a cartoon representation of NCP, PDB ID 6ESF), in which the 153-bp Widom DNA is highly deuterated with 13CH3-methyl labeled at five sites indicated by black circles. The methyl group resonances belonging to the DNA and protein components are shown in red and gray, respectively, with the positions of the m6A methyls aliased (*). The figure was adapted from [65]. (d) Recently developed 19F labeling schemes for nucleic acids. (e) 2D 19F-13C TROSY spectra of 5-fluorouracil (red) modified human hepatitis B virus encapsidation signal epsilon (hHBV ε) element. The figure was adapted from [72].