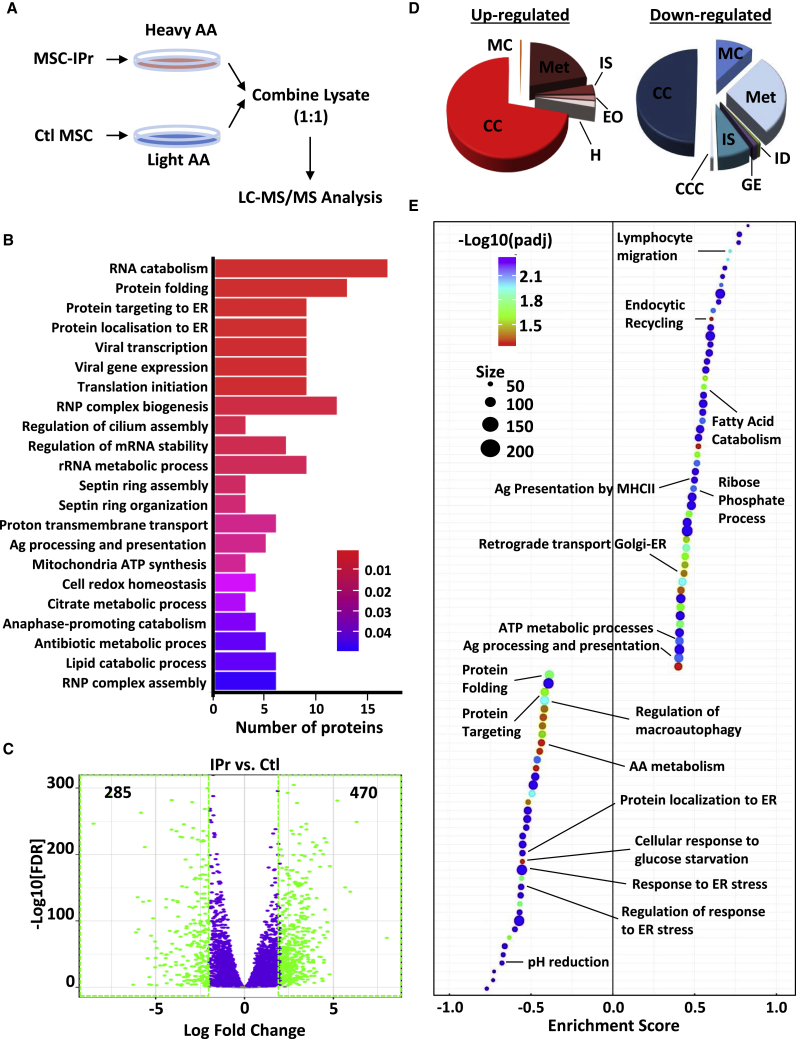
Figure 1.
Functional characterization of MSC-IPr
(A) Representative diagram showing the design of the SILAC experiment.
(B) Bar plot showing the top enriched biological processes based on protein expression analysis from the SILAC experiment (shown in A) and corresponding adjusted p values (false discovery rate [FDR] < 0.05) from a hypergeometric test. The number of proteins contributing to the significance is shown in the x axis.
(C) Volcano plot showing the estimated fold changes (x axis) versus the minus log10 of the adjusted p values (y axis) from DESeq analysis. Significant genes with absolute value of log2 fold changes greater or equal to 2 are shown in green.
(D) Major biological processes groups based on gene expression analyses modulated in MSC-IPr in comparison with control (Ctl) MSCs (transduced with GFP-expressing retroviral particles; same backbone used for MSC-IPr). The annotations for this panel are MC (muscle contraction), IS (immune system), EO (extracellular organization), H (hemostasis), CC (cell cycle), Met (metabolism), ID (infectious diseases), GE (gene expression), and CCC (cell-cell communication).
(E) Plot showing the enriched Gene Ontology (GO) biological processes from an unbiased GSEA analysis of the differentially expressed genes between IPr and Ctl RNA-sequencing (RNA-seq) groups. The FDR threshold is set to 0.05. Features are ranked by the enrichment score from the Kolmogorov–Smirnov test (x axis).
See also Figures S1–S3.