Figure 5.
Comparing oxidative phosphorylation in Ctl MSCs versus MSC-IPr
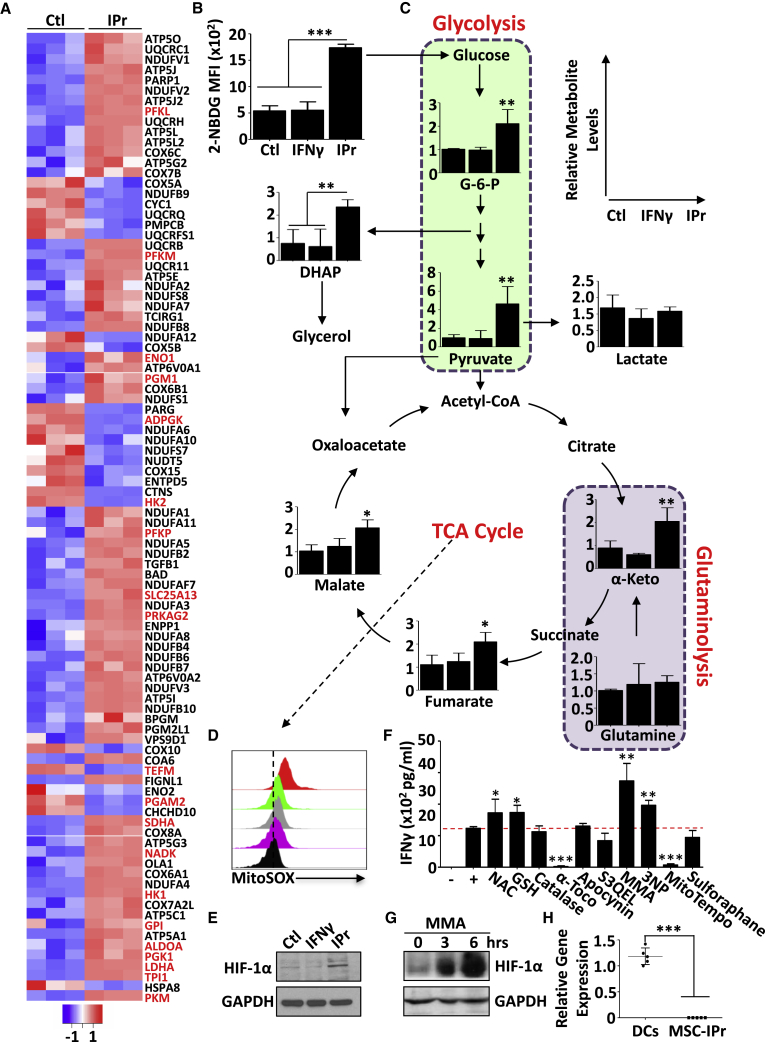
(A) The heatmap represents the Z-scored expression level of the differentially expressed genes from the ATP metabolic process in Ctl MSCs versus MSC-IPr. Upregulated and downregulated genes are highlighted in red and blue, respectively.
(B) Quantification of fluorescent glucose uptake by MSCs.
(C) LC-MS/MS quantification of various glycolysis and TCA metabolites in the three MSC populations.
(D) Representative flow cytometry assessment of superoxide anion by MitoSox. DCs are shown in purple, Ctl MSCs in gray, MSCγ in green, and MSC-IPr in red.
(E) Representative western blot analysis of HIF-1α in Ctl MSCs, MSCγ, and MSC-IPr.
(F) Quantifying IFNγ levels produced by OT-I-derived CD8 T cells co-cultured with OVA-pulsed MSC-IPr treated with various inhibitors targeting ROS or mitochondrial activity.
(G) Representative western blot analysis of HIF-1α in MSC-IPr upon MMA treatment for 3 and 6 h.
(H) Transcript quantification of Sucnr1 by qPCR on DCs and MSC-IPr.
For (B) to (F) and (H), n = 6/group, with ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. Both ANOVA and Student’s t tests were used for this panel. Error bars represent SD. See also Figure S6.