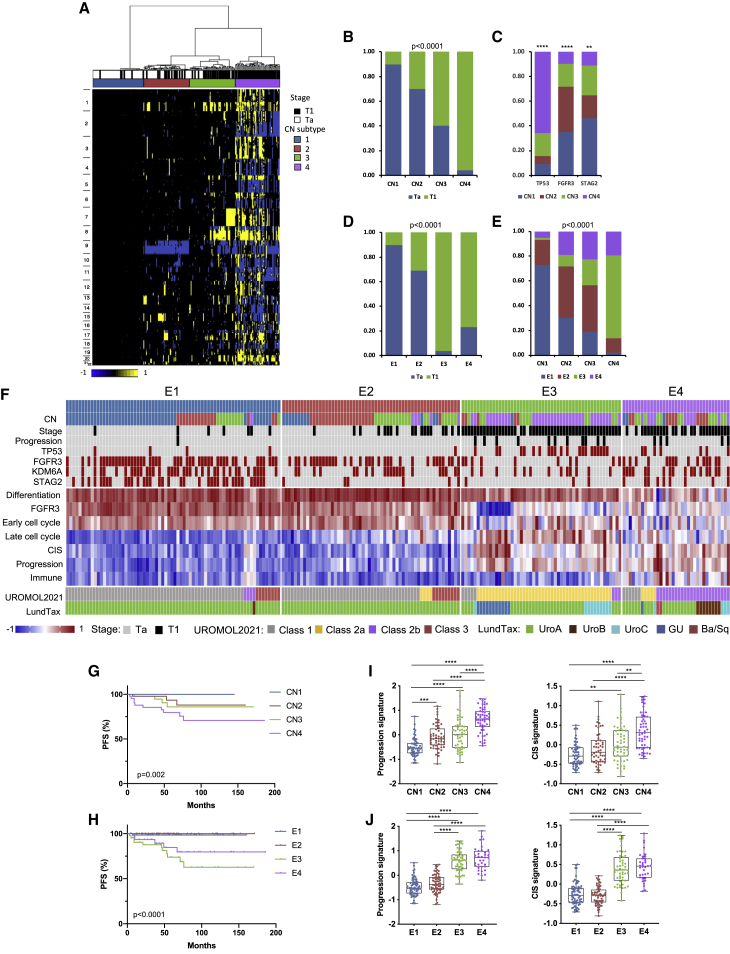
Figure 1.
Combined molecular analysis of stage Ta and T1 NMIBC tumors
(A) CN clusters. Columns, samples; rows, genomic position; yellow, gain; blue, loss. Left: chromosome number. Top: cluster designation and stage.
(B) Relationship of tumor stage to CN subtype.
(C) Distribution of common mutations according to CN subtype.
(D) Relationship of tumor stage to expression subtype.
(E) Relationships of CN and expression subtypes.
(B–E) Chi square test (with Bonferroni correction in B, D, and E).
(F) Top: distribution of CN subtypes, stage, common mutations, and disease progression according to expression subtype. Black, stage T1/progression; red, mutation present. Center: gene expression signatures (standardized Z scores) with differential expression across subtypes (p < 0.0001). Bottom: alignment to UROMOL2021 and LundTax classifications.
(G) Progression-free survival (PFS) stratified according to CN subtype.
(H) PFS according to expression subtype.
(G and H) Log rank analysis.
(I) 12-gene progression risk score7 and weighted CIS score17 in CN subtypes.
(J) 12-gene progression risk score and weighted CIS score in expression subtypes.
(I and J) Kruskal-Wallis test with Dunn’s multiple comparison correction.
Mean, 25th and 75th percentiles, and minimum and maximum values are shown. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01. See also Figures S1 and S2.