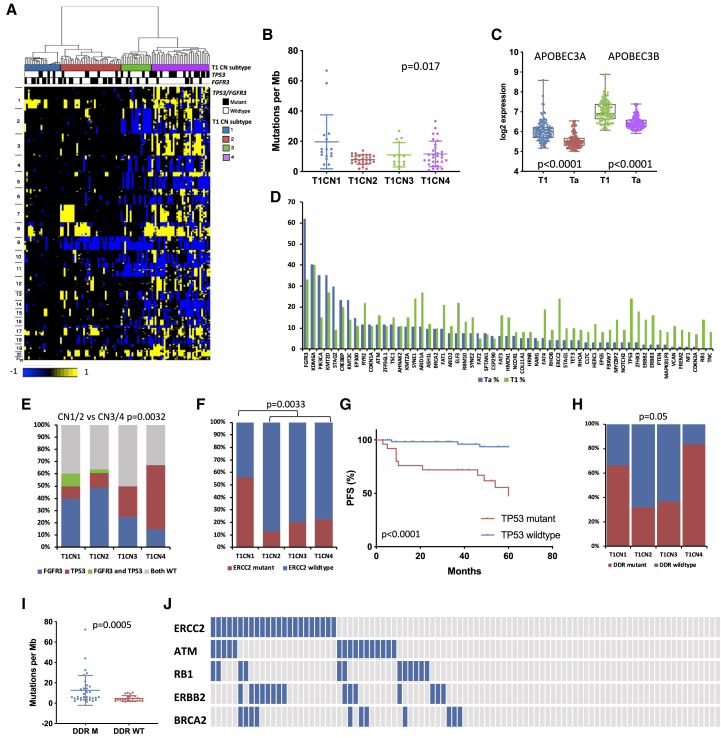
Figure 3.
Independent molecular analysis of stage T1 tumors
(A) CN clusters. Columns, samples; rows, genomic position; yellow, CN gain; blue, CN loss. Left: chromosome number. Top: CN subtype and TP53 and FGFR3 mutation status. Black, mutation present.
(B) TMB as SNVs per megabase according to CN subtype. Kruskal-Wallis test. Bars indicate mean and SD.
(C) Expression of APOBEC3A and APOBEC3B in Ta and T1 tumors.
(D) Mutation frequencies in Ta and T1 tumors for genes mutated in 5% or more of samples in either group.
(E) Distribution of FGFR3 and TP53 mutations in CN subtypes.
(F) Distribution of ERCC2 mutations in CN subtypes. Fisher’s exact text.
(G) PFS according to TP53 mutation status. Log rank analysis.
(H) Mutations in DDR genes analyzed by whole-exome sequencing (any of ERCC2, ATM, RB1, ATR, BRCA2, POLE, FANCC, and CHEK2) according to CN subtype.
(E and H) Chi-square test with Bonferroni correction.
(I) TMB as SNVs per megabase according to mutations in DDR genes (any of ERCC2, ATM, RB1, ATR, BRCA2, POLE, FANCC, and CHEK2) in tumors analyzed by whole-exome sequencing.
(C and I) Mann-Whitney test. Mean, 25th and 75th percentiles, minimum and maximum values are shown.
(J) Relationships of DDR gene mutations. Blue, mutant; gray, wild type.
See also Figure S5.