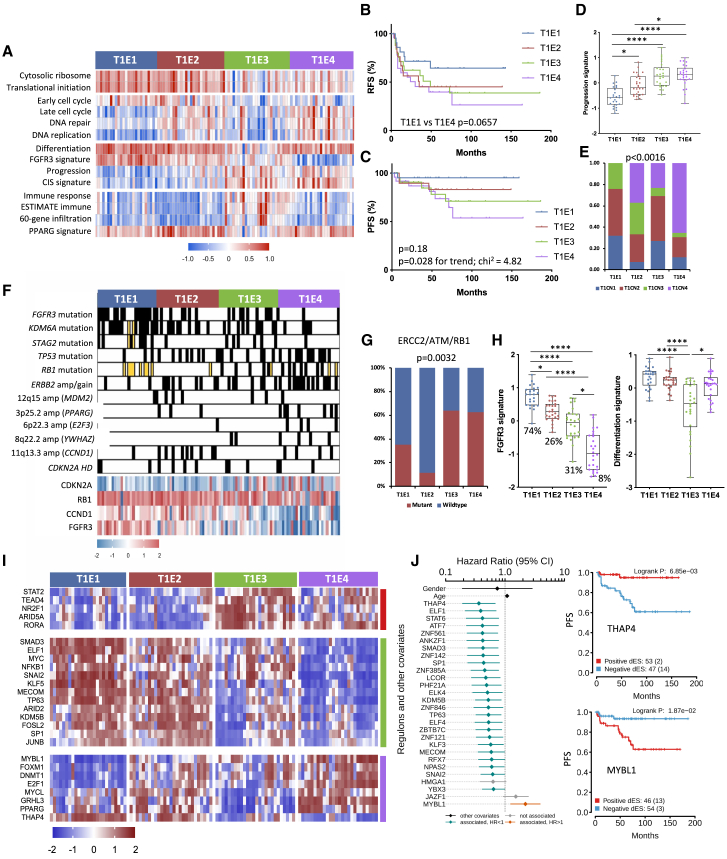
Figure 5.
Features of stage T1 expression subtypes
(A) Heatmaps of Z scores for selected GO categories and expression signatures according to expression subtype. Cytosolic ribosome, GO:22626; Translational initiation, GO:6413; DNA replication, GO:6260; Immune response, GO:6955.
(B) RFS according to expression subtype.
(C) PFS according to expression subtype.
(D) 12-gene progression risk score in expression subtypes.
(E) Relationships of CN and expression subtypes.
(F) Selected CN alterations and gene expression according to expression subtype. Black, amplification or mutation; white, normal or WT; yellow, no data.
(G) ERCC2, ATM, and/or RB1 mutations according to expression subtype.
(E and G) Chi-square test with Bonferroni correction.
(H) FGFR3 signature with mutation frequency and differentiation signature in expression subtypes.
(D and H) Kruskal-Wallis test with Dunn’s multiple comparison correction.
(I) Regulon activity profiles according to expression subtypes. Blocks are color coded according to regulon clusters in Figure S6E.
(J) Left: Cox multivariate regression analysis showing the proportional hazards of each regulon, gender, and age, indicating the contribution of each variable to PFS. Right: Kaplan-Meier plot for PFS, stratified by positive versus negative regulon activity status for THAP4 and MYBL1. BH-adjusted p values.
(B, C, and J right) Log rank analysis.
Mean, 25th, and 75th percentiles, minimum and maximum values are shown. ∗∗∗∗p < 0.0001, ∗p < 0.05. See also Figure S6.