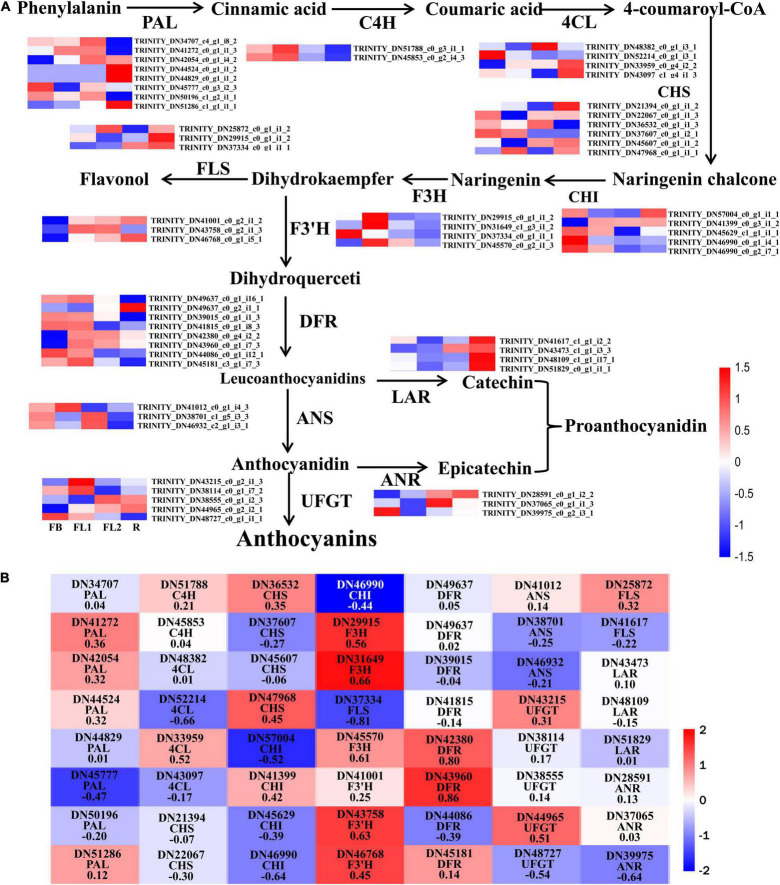
FIGURE 4.
Analysis of DEGs involved in anthocyanin biosynthesis pathway in L. radiata. (A) Anthocyanin biosynthesis pathway and the log2 transformed FPKM values of DEGs associated with structural enzyme genes were used to draw the heatmap. The enzymes include 4-coumarateCoA ligase (4CL), phenylalanine ammonia lyase (PAL), chalcone synthase (CHS), flavone 3-hydroxylase (F3H), chalcone isomerase (CHI), flavonoid 3′-hydroxylase (F3′H), dihydroflavonol reductase (DFR), flavonol synthase (FLS), UDP-flavonoid glucosyl transferase (UFGT), anthocyanidin reductase (ANR), and leucoanthocyanidin reductase (LAR). FB, floral bud stage; FL1, partially opening flower stage; FL2, fully opened flower stage, and R, senescent flower stage. Color gradients comprise red, white, and blue, representing genes that were upregulated, not regulated, as well as downregulated, respectively. (B) The heatmap analysis of all DEGs in anthocyanin biosynthesis pathway according to the FPKM value.