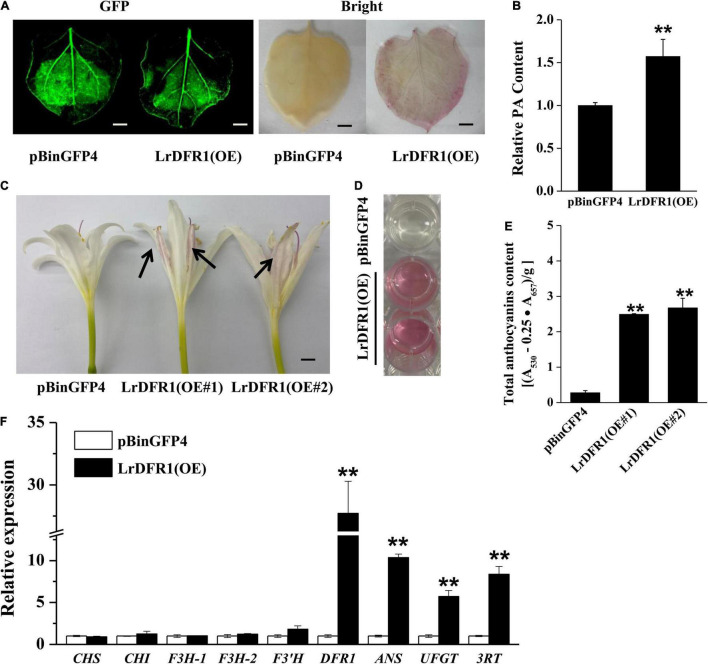
FIGURE 8.
Overexpression of LrDFR1 promotes anthocyanin and proanthocyanidin biosynthesis. (A) Proanthocyanidin staining and (B) relative proanthocyanidin (PA) levels in transiently transformed tobacco leaves (pBinGFP4: empty vector controls; LrDFR1-OE: LrDFR1-overexpressing leaves). Tobacco leaves were kept in a phytotron at 24°C under constant lighting for 5 days. DMACA was used to stain proanthocyanidin. Every experiment was performed using 8–10 leaves for each genotype. Experiments were conducted in triplicates, and a representative image is shown. Proanthocyanidin levels of empty vector controls were set as the reference to 1. Asterisks represent significant differences between control and LrDFR1-overexpressing leaves (**p < 0.01). Bars = 1 cm. (C) Phenotypes of anthocyanin accumulation. Arrow indicates the transfected petals. (D,E) Relative anthocyanin levels in transiently transformed Lycoris petals (pBinGFP4: empty vector controls; LrDFR1-OE: LrDFR1-overexpressing petals). Lycoris petals were kept in a phytotron at 24°C with a constant light for 5 days. Every experiment was performed using 8–10 petals per genotype. Data are shown as mean ± SD. **p < 0.01. Bar = 0.5 cm. (F) Relative expression levels of endogenous anthocyanin biosynthetic genes in pBinGFP4 (empty vector controls) as well as LrDFR1-overexpressing petals. Expression patterns of early biosynthetic genes (CHS, F3H, CHI, and F3′H) as well as late biosynthetic genes (DFR, UFGT, ANS, and 3RT) in petals were investigated. Asterisks represent significant differences between control and LrDFR1-overexpressing petals (**p < 0.01).