Figure 7.
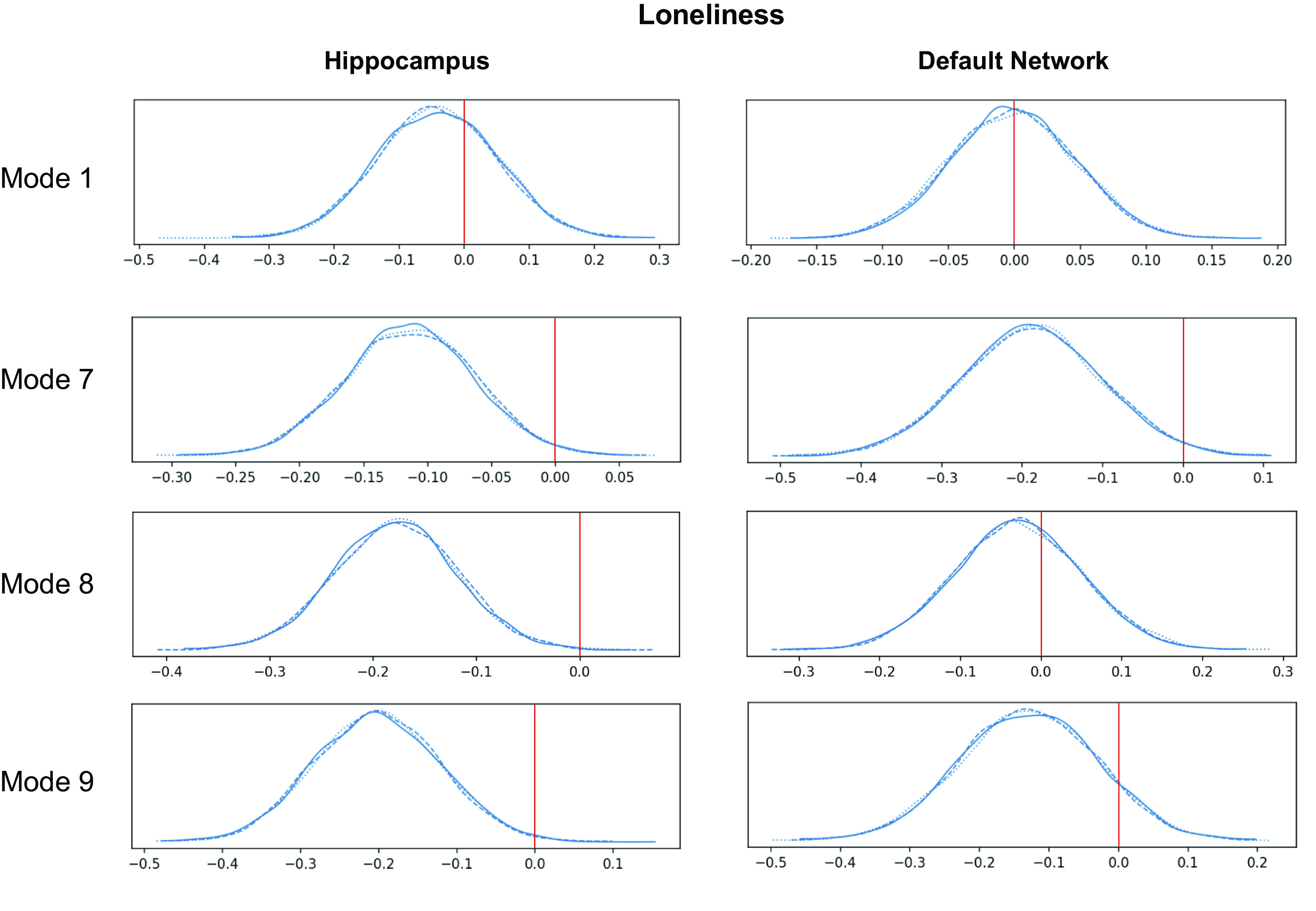
The genetic predisposition for loneliness is associated with the expression of specific hippocampus(HC)-default network (DN) covariation patterns. A polygenic risk score (PRS) analysis was conducted to estimate the subject-specific heritable tendency for loneliness based on genome-wide effects in single-nucleotide polymorphisms. The subject-specific PRS estimates were then regressed against the expressions of each of the modes of structural covariation between the HC and DN. The relevance of the heritability effects was judged based on the posterior parameter distributions inferred by the Bayesian logistic regression model (histograms). The x-axis of each plot represents the β coefficient of the model parameter, and the y-axis of each plot represents the plausibility of each coefficient value. A value of 0 indicates no association between PRS and interindividual mode expression. Overall, loneliness predisposition was related to the expression of modes 7 and 9 on the HC side and to mode 7 on the DN network side. These results demonstrate that specific mode expressions are robustly linked to the predisposition of becoming lonely. The three posterior distribution histograms show convergence across three different Markov chain Monte Carlo runs. Overall, this pinpoints the specific neurobiological signatures in the HC and DN that relate to the heritable components of loneliness.
