Figure 2.
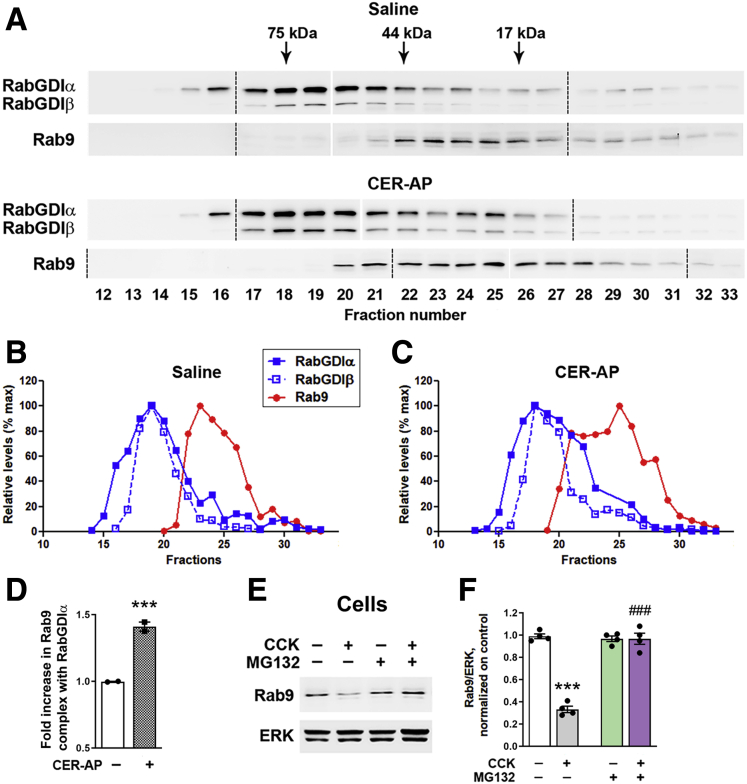
Pancreatitis increases cytosolic Rab9 complex formation with RabGDI and stimulates Rab9 proteasomal degradation. (A–D) Pancreas cytosolic fractions from control animals and those subjected to CER-AP (4 hours) were resolved on a Superdex S200 gel-exclusion column using SMART system (details in Methods). Data are representative of 2 experiments on different animals, with similar results. (A) Eluted fractions were analyzed by IB using antibodies against Rab9 and RabGDIα/β. Samples were run on 3 gels (see full gels in Figure 3), and all the IBs were developed together. Dashed lines denote the first lane in each gel; narrow white space indicates omitted lanes in which protein size ladder was run (see Figure 3). (B and C) Band intensities of Rab9 and RabGDI in each fraction were densitometrically quantified and presented as % of maximal for each protein. (D) Amount of Rab9 in complex with RabGDIα was quantified as a ratio of sum of Rab9 band intensities co-fractionated with RabGDIα to that of total Rab9 (in all fractions 12–33) and presented relative to saline control. In the control (saline-treated) group, the bulk of Rab9 complex with RabGDIα eluted in fractions 22–24; in CER-AP, in fractions 20–25. (E and F) Acinar cells incubated for 30 minutes with and without the proteasomal inhibitor MG132 (50 μmol/L), followed by 30-minute incubation with and without 100 nmol/L CCK. Rab9 level was measured by IB. Values are mean ± SEM from 3 cell preparations. ∗∗∗P < .001 vs control. ###P < .001 vs CCK alone (no MG132). Significance was determined by 2-tailed Student t test (D) or 1-way ANOVA, followed by Tukey multiple comparisons test (F).