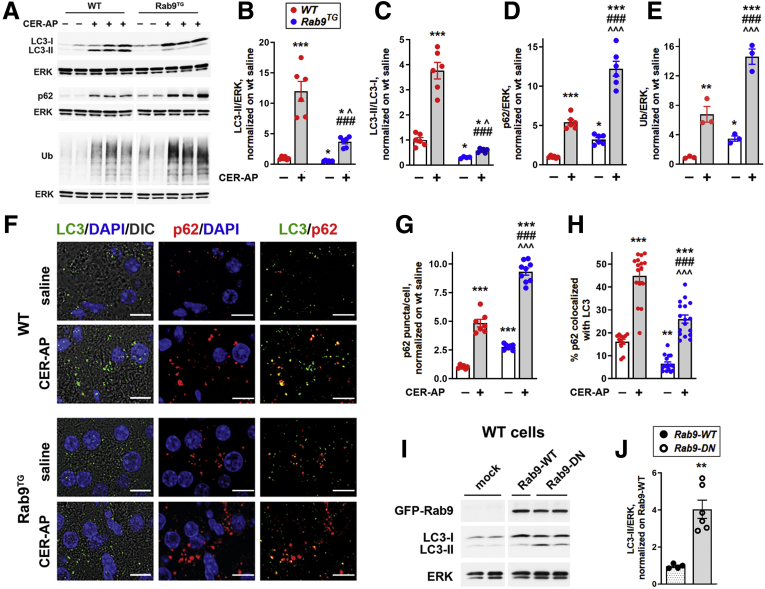
Figure 6.
Rab9 overexpression perturbs canonical/LC3-mediated autophagy in experimental pancreatitis and in control pancreas. (A–E) IB analysis of markers/mediators of canonical autophagy in pancreas of WT and Rab9TG mice subjected to CER-AP (7 hours). Densitometric band intensities for indicated proteins were normalized to ERK in the same sample and further to those in WT control (saline-treated) group. Ub, ubiquitinated proteins. (F–H) IF analysis of pancreatic LC3 and p62/SQSTM1. Scale bars, 10 μm. IF colocalization (H) was quantified with Volocity image analysis software using Manders-Costes coefficients. (I and J) IB analysis and densitometric quantification of LC3-II in WT acinar cells transduced with adenoviral vectors containing GFP-Rab9 (Rab9-WT), GFP-Rab9 dominant-negative S21N mutant (Rab9-DN), or GFP alone (mock), as described in Methods. Values are mean ± SEM from at least 3 animals or cell preparations per group. In (G and H), at least 10 high-power fields per animal were quantified. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 vs WT control (saline) group (B–H) or cells transduced with Rab9-WT vector (J). ###P < .001 vs WT CER-AP. ˆP < .05, ˆˆˆP < .001 vs Rab9TG control (saline) group. Significance was determined by 2-tailed Student t test (J) or 1-way ANOVA, followed by Tukey or Holm-Sidak multiple comparisons test.