Abstract
Mechanisms that regulate nitric oxide synthase enzymes (NOS) are of interest in biology and medicine. Although NOS catalysis relies on domain motions and is activated by calmodulin (CaM) binding, the relationships are unclear. We used single-molecule fluorescence resonance energy transfer (FRET) spectroscopy to elucidate the conformational states distribution and associated conformational fluctuation dynamics of the two NOS electron transfer domains in an FRET dye-labeled endothelial NOS reductase domain (eNOSr) and to understand how CaM affects the dynamics to regulate catalysis by shaping the spatial and temporal conformational behaviors of eNOSr. In addition, we developed and applied a new imaging approach capable of recording three-dimensional FRET efficiency versus time images to characterize the impact on dynamic conformal states of the eNOSr enzyme by the binding of CaM, which identifies clearly that CaM binding generates an extra new open state of eNOSr, resolving more detailed NOS conformational states and their fluctuation dynamics. We identified a new output state that has an extra open conformation that is only populated in the CaM-bound eNOSr. This may reveal the critical role of CaM in triggering NOS activity as it gives conformational flexibility for eNOSr to assume the electron transfer output FMN-heme state. Our results provide a dynamic link to recently reported EM static structure analyses and demonstrate a capable approach in probing and simultaneously analyzing all of the conformational states, their fluctuations, and the fluctuation dynamics for understanding the mechanism of NOS electron transfer, involving electron transfer among FAD, FMN, and heme domains, during nitric oxide synthesis.
Significance
Our results shed light on how CaM and nitric oxide synthase enzymes (NOS) function in biological signaling cascades by revealing how their interaction regulates the conformational behavior of eNOS, which in turn enables the electron transfer reactions that are required to support NO synthesis. Analyzing the conformational and functional domain motions of eNOS activated by CaM binding, we were able to identify that CaM binding generates an extra new open state of eNOSr, a third intermediate, likely critical for electron transfer in the NOS catalytic activities. Our results provide a dynamic link to recently reported EM static structures and demonstrate a capable approach in probing and simultaneously analyzing all of the enzymatic conformational states, their fluctuations, and the fluctuation dynamics.
Introduction
Nitric oxide synthase (NOS) enzymes (1, 2, 3) synthesize nitric oxide (NO) by using a complex assembly of domains and cofactors to convert L-arginine to L-citrulline and NO, via the intermediate N-hydroxy-L-arginine. Mammals express three principal NOS isoforms (endothelial, neuronal, and inducible NOS) that are structurally similar and are all active as homodimers. Each monomer is composed of two primary domains: the N-terminal oxygenase domain and the C-terminal reductase domain (Fig. 1). The reductase domain can be further divided into the FAD/NADPH-binding and the FMN-binding subdomains. The FMN subdomain is connected to the oxygenase domain via a polypeptide linker of ∼30 amino acids in length, which contains a helical binding site for calmodulin (CaM).
Figure 1.
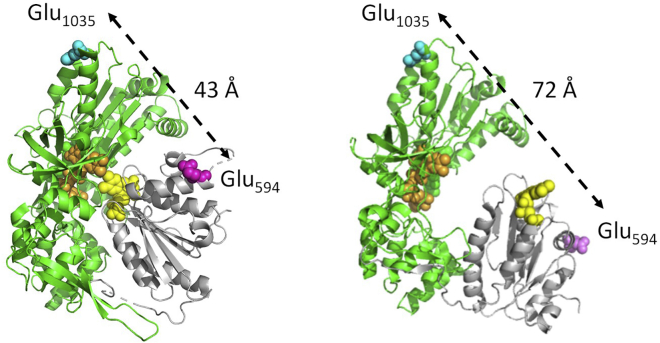
Cartoon structures of CLeNOSr structure depicting the locations of the Glu residues mutated to Cys for Cy3 and Cy5 dye binding. CLnNOSr is shown in a closed conformation (left) and in an open conformation (right). The NADPH/FAD domains are colored green and the FMN domain is gray. The bound FAD and FMN cofactors are colored orange and yellow, respectively, and the two Glu residues are colored blue and pink. The distance between the Glu residues is indicated for either conformation. Models were created based on published crystal structures of neuronal NOSr (PDB: 1TLL) and cytochrome P450 reductase (PDB: 3ES9). PBD, Protein Data Bank ID. To see this figure in color, go online.
The catalytic process involves electron transfer reactions that are correlated with relatively large-scale movement (tens of Ångstrom) of the enzyme domains, in which the reductase domain passes electrons obtained from NADPH through the bound FAD and FMN cofactors and ultimately to the heme cofactor in the oxygenase domain of the opposite monomer (3, 4, 5, 6). Electron transfer is initiated when NADPH reduces FAD by two electrons in the form of a hydride transfer. FMN accepts single electrons through a direct interaction between the FAD/NADPH and FMN subdomains (3, 4, 5, 6). The electron transfers into and out of the FMN domain are the key steps for catalysis, and they appear to rely on the FMN domain cycling between electron-accepting and electron-donating conformational states (7,8). In this model, the FMN domain is suggested to be highly dynamic and flexible due to connecting hinges that allow it to alternate between its electron-accepting (FAD → FMN) or closed conformation and electron-donating (FMN → heme) or open conformation (7,9, 10, 11, 12, 13, 14, 15) In the electron-donating open conformation, the FMN domain has moved away from the FAD/NADPH domain to expose its bound FMN cofactor so that it may transfer electrons to the NOS oxygenase domain or an external acceptor protein such as cytochrome c. In this way, the reductase domain structure cycles between closed and open conformations to deliver electrons, according to a conformational equilibrium that determines the movements and thus the electron flux capacity of the FMN domain (5,7,11,13,14,16).
The CaM binding domain that is located on the peptide linker downstream from the FMN domain also provides an important layer of regulation (3, 4, 5,17). CaM binding to NOS enzymes increases electron transfer from NADPH through the reductase domain and also triggers electron transfer from the FMN domain to the NOS heme as is required for NO synthesis (10,11). The ability of CaM, or similar signaling proteins, to regulate electron transfer reactions in enzymes is unusual, and the mechanism is a topic of interest and intensive study. It has long been known that CaM binding alters NOSr structure to populate a more open conformation (18,19). Recent equilibrium studies have detected a buildup of between two to four discreet conformational populations in NOS enzymes and in related flavoproteins, and in some cases, have also estimated the average distances between the bound FAD and FMN cofactors in the different species (6,16,17,20), and furthermore, have confirmed that CaM shifts the NOS population distribution toward more open conformations (13,15,21), and may enable a transient complex to form between the oxygenase domain-FMN domain-CaM due to the CaM binding (22). Recent structural studies suggested CaM binding induces flexibility in the reductase domain that causes the FMN subdomain to be released and undergo a large rotation for exposure to the oxygenase domain (4,5,14,23,24). However, such ensemble-averaged results about conformational states are not enough to explain how electrons transfer through these enzymes, or how CaM increases the electron flux in NOS, because of an absence of correlated conformational dynamics information. To address this, we are utilizing single-molecule fluorescence energy resonance transfer (FRET) spectroscopy to study how CaM binding affects the conformational states and the domain movement dynamics in NOS enzymes.
During catalysis by full-length NOS enzymes the FMN domain must interact with the oxygenase domain after CaM binds to accomplish electron transfer to the heme, and this interaction of the FMN domain with the oxygenase domain may perturb the overall reductase domain conformational equilibrium to some degree. However, in this work, we utilize a truncated endothelial NOS protein (eNOSr) consisting of the reductase and CaM binding domain of the full-length enzyme to examine how dynamical properties change upon the addition of CaM. There are several reasons for using the eNOSr protein rather than the full-length eNOS enzyme. First, previous reports by our group (19,25,26) and others (6,27) have indicated that the ensemble conformational behaviors of NOSr proteins versus the full-length NOS are similar in response to CaM binding. This has been documented based on how CaM binding increases the electron flux through NOS to cytochrome c, increases NOS flavin fluorescence emission intensity, and changes the NOS FMNH semiquinone magnetic interaction with the NOS FADH hydroquinone or with large paramagnetic ions like dysprosium in solution. This means that the attached oxygenase domain does not fundamentally change the effect of how CaM binding impacts the reductase domain conformational behavior. Secondly, there are practical problems with carrying out the single-molecule studies with the full-length NOS enzyme due to the full-length NOS enzyme existing in functional form as a homodimer. That would complicate our single-molecule measures because we would be observing the relative positions of two FAD-FMN domain FRET pairs simultaneously within a single dimeric NOS molecule. Moreover, because the full-length NOS enzymes contain heme in their oxygenase domains, the variable ability of the bound heme to quench the FRET dye fluorescence would confound the FRET fluorescence signals that we are trying to monitor based on the relative positions of our FRET dyes within the reductase domain. Thirdly, for single-molecule experiments we have to dilute the protein solution to create the single-molecule grid which may cause the eNOS dimer to dissociate, and then any influence of the oxygenase domain interactions on the FMN-FAD equilibrium becomes moot because in NOS enzymes the FMN domain of one subunit only interacts productively for electron transfer with the oxygenase domain of the partner subunit in a functional heterodimer, i.e., any intrasubunit FMN-oxygenase domain interactions occurring in a monomer are nonphysiologic.
Our initial study (28) with neuronal NOSr (nNOSr) indicated that CaM binding alters its conformational behaviors in three ways: It changes the distance distribution between the nNOSr domains, shortens the lifetimes of the individual conformational states, and instills conformational discipline by greatly narrowing the distributions of the conformational states and fluctuation rates. This helped reveal how CaM promotes catalysis by shaping the physical and temporal conformational behaviors of nNOS, which is important for a fundamental understanding electron transfer dynamics in a protein-redox respirational system (28).
Despite their structural similarity, the three NOS isoforms each have unique catalytic profiles (3,29). The differences are greatest between the endothelial NOS (eNOS) and neuronal NOS (nNOS) isoforms; eNOS has a 6-fold lower NO synthesis activity and a 6–16-fold lower cytochrome c reductase activity than nNOS (13,30,31). Studies suggest that these differences are primarily due to structural differences within their reductase domains (13,30,32,33). To better understand the differences between nNOS and eNOS, we investigated how CaM binding affects domain conformation and movement dynamics in eNOSr by using single-molecule FRET spectroscopy.
Materials and methods
Sample preparation and characterization
Materials and preparation of the enzyme solution
CaM bovine (C4874–10KU, lyophilized powder, ≥98%) and β-nicotinamide adenine dinucleotide phosphate sodium salt hydrate (NADP; N0505, ≥98%) were purchased from Sigma-Aldrich (St Louis, MO, USA). 6-OH-FAD was a gift from David Ballou (University of Michigan, Ann Arbor, MI). 6-OH FMN was generated from 6-OH-FAD using snake venom phosphodiesterase I and then purified as described (34) and the conversion and purity were checked in HPLC. The buffer solution used for experiments was 40 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) with 10% (vol/vol) glycerol and 150 mM NaCl, pH 7.6.
To overcome a dilution-based dissociation of FMN from the eNOSr, we added 6-OH-FMN, which has much less fluorescence than FMN but still binds within eNOSred and supports its catalysis. The 6-OH-FMN enabled us to avoid a prohibitively high fluorescence background that otherwise became present if FMN was instead added to the diluted enzyme solution. Experimental solutions contained 5 nM dye-labeled eNOSr, 10 μM NADP, and 2 μM 6-OH-FMN in the buffer. In some cases, 10 μM CaM and 400 μM CaCl2 were also added.
Preparation of samples for FRET measurements
The samples for single-molecule spectroscopic measurements were prepared by mixing 4 μL of enzyme solution described above, with 6 μL of oxygen scavenger solution that contained 0.8% D-glucose, 1 mg/mL glucose oxidase, 0.04 mg/mL catalase, and 1 mM of 6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (Trolox) and is added to prevent FRET dye molecule photobleaching or blinking due to forming triplet state (35,36), and with 10 μL of 1% agarose that had been heated just above its gel-transition temperature (26°C). The mixture was then placed between two glass coverslips to form a sandwich. The final concentration of eNOSred is ∼1 nM. Once the solution cools it forms a gel containing dispersed single-enzyme molecules that are entrapped in chambers large enough to allow the enzyme molecule to freely rotate, and to allow small molecules to diffuse freely. Our previous anisotropy study has demonstrated that confining single enzyme molecules in the agarose gel in this way has no perturbation on the enzyme motions. In our experiments, 1 nM eNOSr with oxygen scavenger solution was sandwiched between two clean glass coverslips in 1% agarose gel (in 99% water). In the 1% agarose gel, single-eNOSr-enzyme molecules can rotate freely (37, 38, 39).
Single-molecule imaging and FRET measurements
We used the single-molecule photon-stamping approach to record the single-molecule FRET fluctuation time trajectories photon-by-photon for both the donor and acceptor simultaneously (37,39, 40, 41). A detailed description of the experimental setup has been published in our previous publication (37,39, 40, 41). Briefly, the fluorescence images and photon-counting trajectories were acquired with an inverted confocal microscope (Axiovert 200; Zeiss, Oberkochen, Germany). The excitation laser (520-nm 80-MHz pulse laser; Coherent, Santa Clarita, CA, USA) confocal beam was reflected by a dichroic beam splitter (z532rdc; Chroma Technology, Bellows Falls, VT, USA) and was focused by a high-numerical-aperture objective (1.3 NA, 100×; Zeiss) on the sample at a diffraction-limited spot of ∼300-nm diameter. To obtain the fluorescence images and intensity trajectories, the emission signal was split by using a dichroic beam splitter (640dcxr) into two-color beams centered at 570- and 670-nm representing Cy3 and Cy5 emissions, respectively. The two channels of the signal were detected by a pair of Si avalanche photodiode single-photon-counting modules (SPCM-AQR-16; Perkin-Elmer Optoelectronics, Wiesbaden, Germany). Typical images (10 × 10 μm) were acquired by continuously raster-scanning the sample over the laser focus with a scanning speed of 10 ms/pixel, with each image of 100 pixels × 100 pixels. The fluorescence intensity trajectories of the donor (Cy3) and acceptor (Cy5) were recorded by a two-channel Picoharp 300 (PicoQuant, Berlin, Germany) photon-stamping setup. The arrival of each detected photon from a donor (Cy3) and acceptor (Cy5) fluorescence was sampled and treated into trajectory with 10-ms binning time and the intensity trajectories were typically collected for 60 s. We resolve the conformational changes in FRET pair distance changes from millisecond timescale data analysis in millisecond time bin photon-counting time trajectory analysis, and the dipole-dipole-orientation-regulated FRET efficiency changes occur at a much shorter timescale of nanoseconds to picoseconds, should be averaged out in millisecond time bin.
Generation of Cys-light eNOSr
We utilized mass spectrometry to identify maleimide dye-reactive Cys residues that are present in bovine eNOSr, and then utilized site-directed mutagenesis to convert these to Ser, thus generating a “Cys-lite” version of eNOSr that displayed only residual reactivity toward the FRET dyes. Based on the crystal structures of the homologous proteins, neuronal NOSr (7) and cytochrome P450 reductase (20,42,43), we then substituted Cys residues for surface Glu residues 594 and 1035 on the enzyme’s FMN and FAD/NADPH domains, respectively, that are usefully positioned for our FRET studies, creating the E594C/E1035C Cys-lite eNOSr (henceforth referred to as CLeNOSr).
Expression and purification of eNOSr and CLeNOSr proteins
eNOSr and CLeNOSr (13,44) protein expression was induced at room temperature over one or 2 days in Escherichia coli BL21(DE3) as previously described (13,44,45). The proteins were purified by sequential chromatography on a 2′,5′-ADP Sepharose affinity column and CaM-Sepharose resin (Millipore Sigma, St Louis, MO, USA) as reported (13,44,45). Purity of each protein was assessed by SDS-PAGE and spectral analysis. The concentration was determined by using an extinction coefficient of 22,900 M−1 cm−1 at 457 nm for the fully oxidized form (13,44,45).
Steady-state cytochrome c reduction assay
The cytochrome c reductase activity was determined at room temperature by monitoring the absorbance increase at 550 nm and using a difference extinction coefficient ε550 = 21 mM−1 cm−1 as described previously (13,42,44).
Estimation of the CLeNOSr conformational Keq
The open/closed conformational Keq was determined by following the reduction of excess cytochrome c by fully reduced CLeNOSr (CaM-free or CaM-bound) in a stopped-flow instrument at 10°C as described previously (13,33,44,45). The CLeNOSr (10–12 μM) in 40 mM EPPS buffer (pH 7.6) with 10% glycerol, and 150 mM NaCl containing EDTA (2 mM) and 200 μM NADPH ware fully reduced by titrating it with sodium dithionite solution under anaerobic conditions. This solution was rapid-mixed in the stopped-flow instrument with an anaerobic solution of cytochrome c (100 μM) while monitoring the absorbance changes at 550 nm. The magnitude of the absorbance changes representing the fast-reacting and slow-reacting phases of the reaction during the reduction of the first equivalent of cytochrome c was determined and used to estimate the conformational keq (42,44,45).
We note that we have traditionally carried out our pre-steady-state kinetic measures on NOS enzyme species at 10°C to slow down the reactions and thus enable more accurate data collection and comparisons. We have not measured the temperature effects on the open-closed conformational equilibrium for the NOSr enzymes in the presence and absence of bound CaM. However, steady-state measures of eNOSr electron flux to cytochrome c have been made at room temperature versus 10°C (13). These measures indicate that the impact of CaM binding on the conformational equilibrium is somewhat reduced at the lower temperature, but its binding still has the same overall effect on the eNOSr, which causes a conformational opening at both temperatures.
Results and discussion
Ensemble-averaged experiments to validate the CLeNOSr
CLeNOSr displayed near-normal cytochrome c reductase catalytic activity both in its native and CaM-bound states (Table 1). This indicates that the substitutions we incorporated into eNOSr do not significantly change its catalytic behavior or response to CaM.
Table 1.
Steady-state (min−1) cytochrome c reductase activity of each eNOSr protein in the CaM-free or CaM-bound states
| Protein | –CaM | +CaM |
|---|---|---|
| Wild-type eNOSr | 110 ± 9 | 450 ± 41 |
| CLeNOSr | 145 ± 14 | 561 ± 22 |
Reactions were run at 25°C as described in Materials and methods.
We next carried out ensemble experiments for verifying the conformational changes of CLeNOSr under CaM binding and activation. The incorporated Cys594 and Cys1035 surface residues are located outside of the predicted FMN and FAD/NADPH domain-domain interface in CLeNOSr and are also located distant from the NOS CaM-binding helix (46) and are expected to fluctuate within a distance range of ∼40–70 Å for the closed and maximally open conformational states, respectively, based on structural similarities to the open and closed states of the related enzyme cytochrome P450 reductase (Fig. 1). This should allow for monitoring of conformational changes by FRET of the Cy3-Cy5 dye pair, using a corrected Forster radius of Ro, 45 Å (28).
The CLeNOSr could be double-labeled by the Cy3 and Cy5 maleimide dyes. The double-labeled protein displayed a strong FRET fluorescence signal, as judged compared with the signal obtained for an equal mixture of CLeNOSr proteins that were singly-labeled with either Cy3 or Cy5, which could not engage in FRET (Document S1). CaM binding to the doubly labeled CLeNOSr protein caused an observable loss of FRET signal intensity, as indicated by the gain in donor fluorescence signal at 525 nm (Document S1). This means that CaM binding caused the Cy3 and Cy5 dyes to move further apart on average, consistent with CaM-bound CLeNOSr favoring more open conformational states (18,19). Single turnover measures of cytochrome c reduction by the fully reduced CLeNOSr as monitored in a stopped-flow instrument independently confirmed that CaM binding shifts the conformational equilibrium of CleNOSr to favor more open states, as indicated by CaM causing a gain in the open/closed conformational Keq of CLeNOSr (Table 2). Overall, the ensemble data collected for CLeNOSr show that it maintains a near-normal response to CaM regarding activation of catalysis, and when labeled with Cy3/Cy5 dyes can indicate by FRET the conformational change caused by CaM binding. Thus, CLeNOSr was judged to be a useful probe for single-molecule studies.
Table 2.
Effect of CaM binding on the conformational equilibrium (Keq) of fully reduced CLeNOSr
| Condition | Fraction open | Fraction closed | Keq | |
|---|---|---|---|---|
| CLeNOSr | –CaM | 0.53 ± 0.025 | 0.47 ± .025 | 1.1 ± 0.12 |
| +CaM | 0.65 ± 0.03 | 0.35 ± 0.03 | 1.9 ± 0.22 |
An excess of cytochrome c was mixed with the fully reduced CLeNOSr protein in a stopped-flow instrument at 10°C, and the rate of cytochrome c reduction was followed at 550 nm to estimate the fraction of CLeNOSr in the open (reactive) and closed (unreactive) conformations, as described under “Materials and methods.” Values are an average of 6–7 individual mixings done under identical conditions, using two different protein preparations for each mutant.
Single-molecule spectroscopic studies of enzymatic conformational dynamics
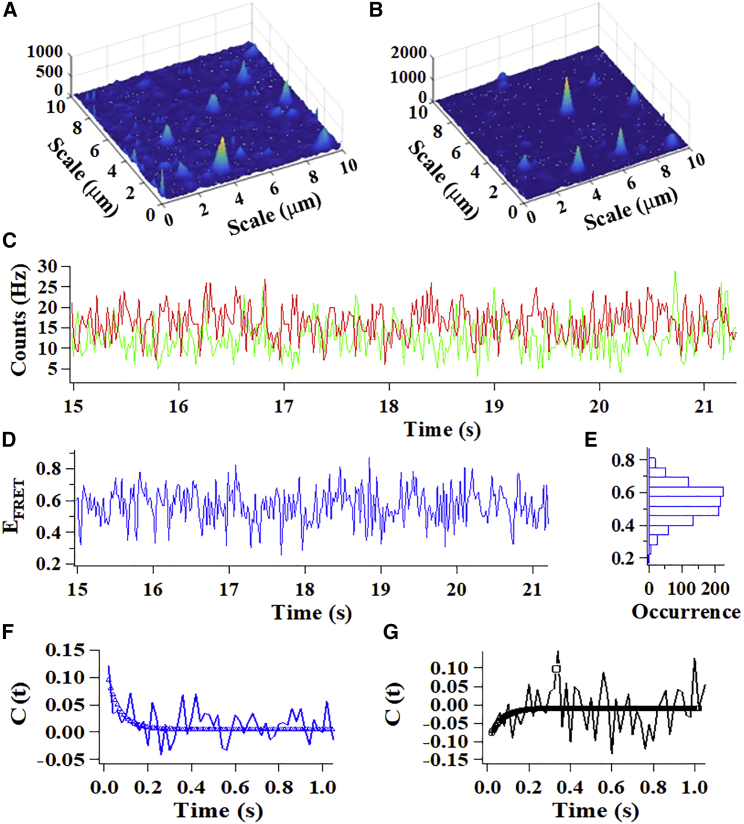
We next performed single-molecule experimental investigations of CLeNOSr to study its conformational dynamics and to probe the mechanism of how CaM binding regulates CLeNOSr electron transfer processes that are fundamental to its redox functions. Fig. 2, A and B shows typical donor-acceptor (D-A) two-channel fluorescence images collected for single molecules of Cy3-Cy5-labeled CLeNOSr within a 10 × 10-μm laser confocal scanning area in the agarose gel. Each feature in the images is attributed to a single enzyme molecule; the intensity variation between the molecules is due to FRET and the different longitudinal positions in the light focal depths. Protein conformational changes that cause changes in FRET D-A distance result in coincident and opposite, anticorrelated donor and acceptor fluorescence signal fluctuations (donor decreases, acceptor increases, and vice versa). Fig. 2 C shows the intensity versus time trajectories of the Cy3 donor (Id(t), green)- and Cy5 acceptor (Ia(t), red)-labeled CLeNOSr under the CaM-free condition. Document S1 D shows the FRET efficiency versus time (EFRET∼t) trajectory, calculated as EFRET = Ia/(Id + Ia) from Fig. 2 C. The EFRET∼t trajectory reflects the protein conformational motion dynamics, indicating that the Cy3-Cy5-labeled domains of CLeNOSr fluctuate at a certain rate, which provides a statistical result of the protein conformational states. A histogram (Fig. 2 E) deduced from the EFRET∼t data of a single CLeNOSr molecule reflects the distribution of molecular conformations it adopted over the 0- to 21-s recording period and shows that they arrange according to a normal distribution, with the most populated conformational states for this molecule corresponding to an EFRET of 0.54.
Figure 2.
Single-molecule fluorescence images (10 × 10 μm) of Cy3/Cy5-labeled CLeNOSr, the emission is from the FRET dyes. (A) Donor. (B) Acceptor. (C) A portion of a single-molecule intensity versus time trajectory: green represents donor, and red represents acceptor. (D) Single-molecule FRET efficiency versus time trajectory. (E) The distribution of the FRET efficiencies. (F) Autocorrelation of FRET efficiency versus time trajectory. (G) Cross-correlation of donor and acceptor from intensity versus time trajectory. To see this figure in color, go online.
Typically, single-molecule D-A signal fluctuation involves not only FRET-related fluctuation, which shows an anticorrelated relation between the D-A signal fluctuations; but also noncorrelated thermal-related random fluctuation, and measurement noise. To extract the conformational dynamics information from the intensity versus time trajectories of the Cy3 donor (Id(t), green)- and Cy5 acceptor (Ia(t), red)-labeled CLeNOSr, we applied autocorrelation and cross-correlation function analysis to characterize the fluctuations due to the FRET energy transfer process. Fig. 2 F shows the autocorrelation functions of the FRET signals, and Fig. 2 G shows the cross-correlation function between donor and acceptor signals. All of the correlation functions can be fit with the same decay rate constant within a standard deviation, which confirms that both the donor and acceptor signal fluctuations arise from the same origin, namely the single-molecule protein conformational fluctuations.
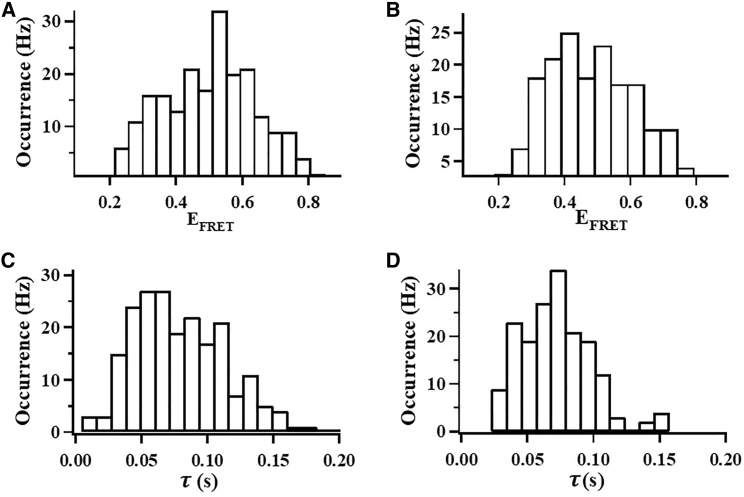
To study how CaM binding impacts the conformational distribution of CLeNOSr, we analyzed single molecules of its CaM-free or CaM-bound forms as described in Fig. 2, and determined each molecule’s mean EFRET-value (as in Fig. 2, D and E), and then plotted the distributions of these means. The histograms (Fig. 3, A and B) show that the CaM-free and the CaM-bound CLeNOSr molecules both populate many conformations that have a range of EFRET-values from 0.23 to 0.78. Notably, the mean FRET efficiency is ∼0.5 without CaM binding and is 0.48 with CaM binding. Based on the Forster radius of Cy3 and Cy5 and related considerations, this corresponds to an increase in the Cy3–Cy5 label mean distance from 4.5 to 4.6 nm upon CaM binding to CLeNOSr, assuming R0 is 4.5 nm. It is important to note that these mean distance values are a statistic result from measures of dynamic structural fluctuations, and the range of structural fluctuation distances that are adopted by any individual molecule is more extensive. The mean values can also fluctuate depending on the dwell times of individual states over the course of the collection time (for example, shorter-lived conformations may appear more times during the collection period). Nevertheless, our study provides a dynamics-based demonstration that under physiologic conditions, CaM binding mildly shifts the conformational distribution of CLeNOSr toward more open states, consistent with results from ensemble-based studies (3,6,13,16). This conclusion is consistent with the extent of FRET loss that we observed upon CaM binding in the ensemble measurement in Document S1, and matches the extent to which CaM alters the open-closed conformational equilibrium of a fully reduced CLeNOSr that we determined by ensemble measurement using an established stopped-flow spectroscopic technique (42,44,45) (Table 2).
Figure 3.
(Upper) Histograms of the FRET efficiency distribution for Cy3/Cy5-labeled CLeNOSr from the EFRET versus time trajectories (each occurrence is the mean value from histograms as in Fig. 2E) without (A) or with (B) CaM. (Lower) Histograms of the delay time distribution from the autocorrelation of the intensity versus time trajectories without (C) or with (D) CaM.
Single-molecule data can also inform on the conformational fluctuation dynamics of CLeNOSr and on how CaM binding impacts the dynamics, which is an important facet that determines electron flux through the enzyme. Fig. 3, C and D contain histograms of the fluctuation correlation time τ related to the dwell times that we measured for single molecules in any given conformational state, as derived from the autocorrelation functions of the single-molecule FRET intensity time trajectories. For CaM-free CLeNOSr, the conformational fluctuation correlation times τ were distributed broadly across 20–150 ms, with an average value of ∼80 ms. These data demonstrate that CaM-free CLeNOSr molecules adopt a range of shorter- and longer-lived conformational states in solution whose lifetimes vary over 10-fold. CaM binding to CLeNOSr caused an apparent change in the conformational fluctuation correlation times τ (Fig. 3 D). CaM shortened the dwell times and also greatly tightened their distribution, such that the enzyme molecules now displayed fluctuation correlation times within the 20–110 ms time window, with most having times ∼70 ms (Fig. 3 D). In general, these changes were also observed when CaM binds to CLnNOSr (28).
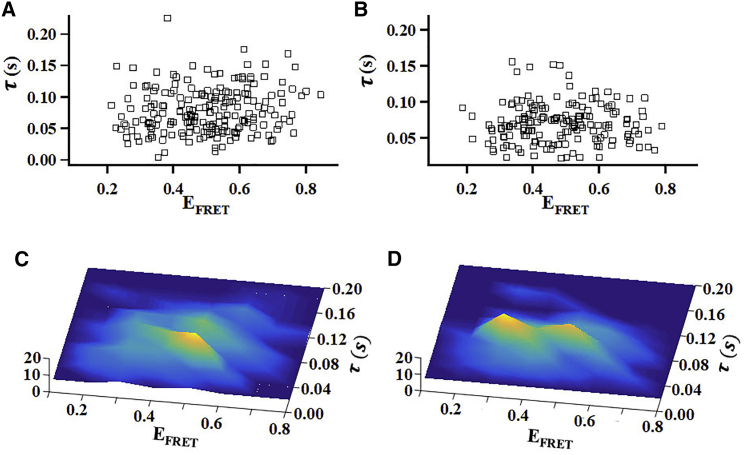
To further illustrate how CaM binding impacts the conformational dynamics of CLeNOSr, we plotted the pairs of conformational fluctuation correlation times τ- and EFRET-values that we observed from many individual CLeNOSr molecules, without or with bound CaM (Fig. 4, A and B) and plotted them as the three-dimensional (3D) topographical distributions (Fig. 4, C and D). Several interesting and fundamental concepts emerge from the graphic analysis: First, with CaM binding, besides the peak at the position of EFRET at ∼0.5, there is another new peak appearing at the position of EFRET at ∼0.3, indicating the CaM binding caused a new more open conformation to populate in the molecular distributions. Secondly, CaM binding reduced or eliminated the most long-lived conformations of CLeNOSr that are otherwise populated under the CaM-free condition, and whose presence may be expected to retard steady-state electron transfer during catalysis. Third, the graphs show that there is no strong correlation apparent between a CLeNOSr molecule’s conformational state (EFRET) and its conformational dwell time τ in either the CaM-free or CaM-bound condition. This result indicates that longer-lived states of CLeNOSr do not preferentially populate more closed or more open conformational states, and instead are distributed fairly evenly across the entire range of conformations.
Figure 4.
Impact of CaM binding on the distribution of single Cy3/Cy5-labeled CLeNOSr molecules regarding their conformational fluctuation correlation times (τ) and their associated FRET efficiencies (EFRET). (A and B). (C and D) 3D histograms plotting the molecular pair distributions in (A and B). The heights of the peaks and their colors are proportional to the number of molecules, as indicated by the bars. Here (A and C) are without CaM and (B and D) are with CaM. To see this figure in color, go online.
When compared to our previous results with CLnNOSr, we see that CaM binding affects the conformational states and the domain movement dynamics of CLnNOSr and CLeNOSr in similar ways: It changes the distance distribution between their domains and shortens the lifetimes of the individual conformational states. However, CaM binding causes smaller shifts toward open conformational states and less narrowing of the distributions of the conformational states and fluctuation rates in the CLeNOSr compared to CLnNOSr. Furthermore, the conformational fluctuation rates of CLeNOSr are slower than that of CLnNOSr both with and without CaM binding, consistent with estimates of the conformational fluctuation rates from ensemble measures (3,33).
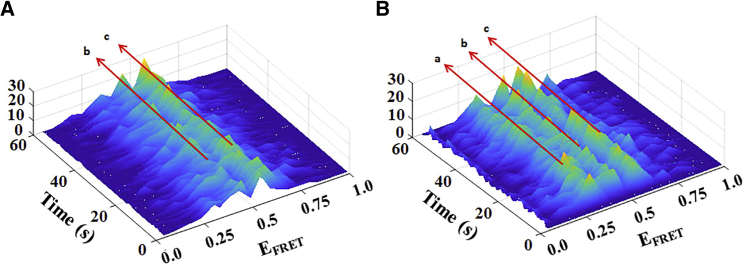
From our single-molecule FRET data, we created histograms of the FRET fluctuation time trajectories to obtain the average FRET efficiencies and thus characterize the conformational distribution, as shown in Fig. 2, D and E. This process averaged out the detailed dynamics information that is present in the FRET fluctuation time trajectory. To avoid losing this conformational dynamics information, and to further understand how CaM binding impacts the conformational behaviors of CLeNOSr, we separated the FRET fluctuation time trajectory into 1-s intervals, then plotted each 1-s distribution pattern versus time to create a 3D image of the FRET histograms versus time. The results in Fig. 5 show that for CaM-free eNOSr, two ranges of FRET peaks dominate with time, located approximately at EFRET 0.4 and EFRET 0.5. However, with the binding of CaM, another additional peak range appears, located approximately at EFRET 0.28, along with the two continuous peak ranges at EFRET 0.4 and FRET 0.5. These results clearly indicate that the CaM-free CLeNOSr favors two conformational states and that CaM binding results in a third more open conformational state being populated at EFRET 0.28. Our observing three predominant conformational states of CaM-bound CLeNOSr corresponds with the three states model proposed by Feng et al. (47, 48, 49). Specifically, the closed conformational state (highest EFRET) may correspond to the input state where the FMN domain is docked to the FAD/NADPH domain, in a position to receive an electron. The most open conformational state corresponds to the output state (lowest EFRET), where the FMN domain is extended away in a position to dock to the oxygenase domain. In this state, the intersubunit FMN−heme IET event could take place. Between the closed conformational state and the most open conformational state, there is an intermediate open conformational state (intermediate EFRET), where the FMN domain is free but not docked and is able to transfer an electron to cytochrome c. In this intermediate conformation, the FMN domain could either go on to transport an electron toward the heme domain or return in its oxidized form to the FAD/NADPH domain to receive a new electron.
Figure 5.
3D histograms plotting single-molecule FRET efficiency (EFRET) distributions of Cy3/Cy5-labeled CLeNOSr with time. The heights of the peaks and their colors are proportional to the occurrence (Hz) of the EFRET, as indicated by the bars. The results indicate two EFRET peaks without CaM (A) and three EFRET peaks with CaM (B). 1) EFRET = 0.28; 2) EFRET = 0.4; 3) EFRET = 0.5. To see this figure in color, go online.
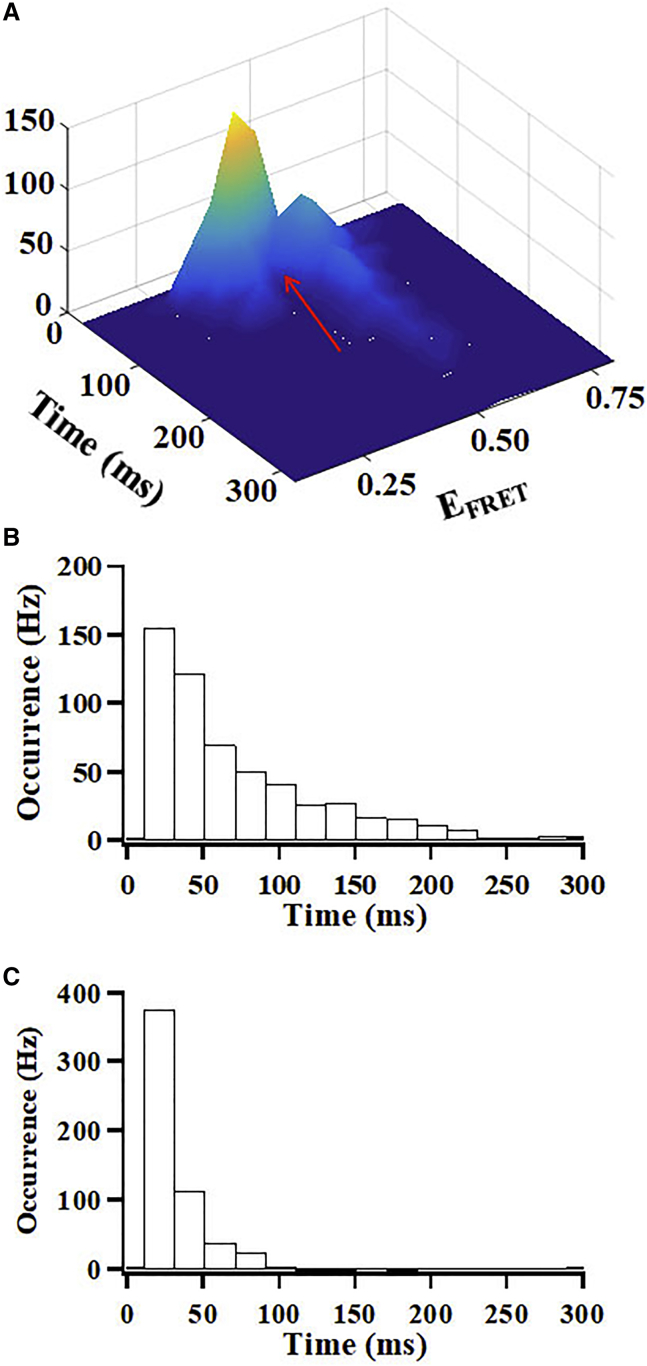
We further analyzed the fluctuation rates and the population probabilities of the conformational states of CLeNOSr without CaM binding. From data in Fig. 5 A, we know that for CaM-free eNOSr there are two conformational state FRET peaks that dominate with time, located approximately at EFRET 0.4 and EFRET 0.5. Accordingly, we set a threshold value (EFRET = 0.45) to separate these two dominant conformational states so we could obtain the CLeNOSr population dwell times at either conformational state. Fig. 6 A shows a 3D histogram plotting single-molecule FRET efficiency (EFRET) distributions with the period of time that molecules dwell at a given EFRET, and indicates the location of the separation threshold (EFRET = 0.45). Fig. 6, B and C show the probability dwell time distributions of the two conformational states. The average dwell time for CaM-free CLeNOSr determined in our single-molecule study is ∼75 ms for molecules populating the closed conformational state, and they display a broad distribution of dwell times. In contrast, the average dwell time for CaM-free CLeNOSr molecules in the open conformational state is 31 ms, and they display a relatively tighter distribution of dwell times. Thus, the closed conformational state is longer-lived than the open conformational state, which corresponds fairly well with our ensemble-based estimate of the conformational equilibrium (Table 2) (13,15). that indicated the CaM-free CLeNOSr favors the closed conformational state. Our current findings show that CaM-free CLeNOSr exhibits a wide range of conformational lifetimes, with the more stable closed conformers likely helping to limit the steady-state electron flux during catalysis.
Figure 6.
(A) 3D histograms plotting single-molecule FRET efficiency (EFRET) distributions of Cy3/Cy5-labeled CLeNOSr versus the periods of time that molecules stay at the given EFRET. The EFRET peaks indicate there are two main conformational states without CaM. The dwell time distributions of the closed state (B) and the open state (C). Red arrow indicates the threshold value for separation of the two states (EFRET = 0.45). To see this figure in color, go online.
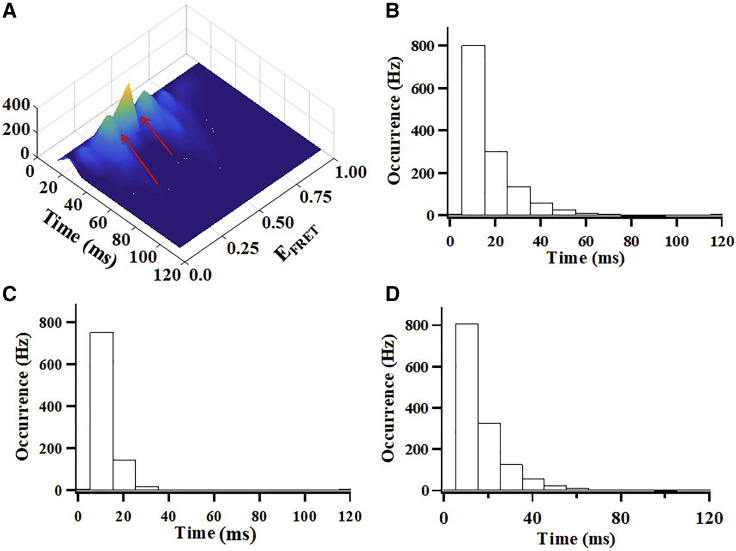
Fig. 5 B indicates that there are three major conformational states populated by the CaM-bound CLeNOSr, located approximately at EFRET 0.28, EFRET 0.4, and FRET 0.5. To study the dwell time distributions of the three conformational states, we set two threshold values (EFRET = 0.35 and EFRET = 0.45) to separate the three conformational states. Fig. 7 A shows the 3D histograms plotting single-molecule FRET efficiency (EFRET) distributions with the period of time that the CaM-bound CLeNOSr molecules dwell at a given EFRET, and indicates the location of the two separation thresholds. Fig. 7, B–D shows the probability distribution of the three conformational states. The results indicate that the dwell times for CaM-bound CLeNOSr are much more tightly distributed than in CaM-free CLeNOSr, and on average are ∼20 ms for the closed state, ∼11 ms for the intermediate conformational state, and ∼18 ms for the open conformational state.
Figure 7.
(A) 3D histograms plotting single-molecule FRET efficiency (EFRET) distributions of CaM-bound Cy3/Cy5-labeled CLeNOSr versus the dwell times that the molecules stay at the given EFRET. The FRET peaks indicate that three major conformational states exist. The dwell time distributions of the closed state (B), intermediate state (C), and the open state (D).
This dwell time analysis is consistent with CLeNOSr populating the intermediate and open conformational states with CaM binding. We propose a model for CaM-mediated regulation of CLeNOSr as shown in Document S2. Without CaM, the FMN domain primarily fluctuates between an intermediate open conformational state and a closed conformational state, and it tends to stay in the closed conformational state longer than in the intermediate open conformational state. CaM binding to CLeNOSr has two effects: It causes CLeNOSr to predominantly populate three conformational states, which are the two previously described plus an additional more open conformational state. It also shortens and equalizes the lifetimes of the closed and open conformational states.
Thus our data reveal that CaM alters CLeNOSr conformational behaviors in three ways: 1) It supports faster conformational fluctuations (i.e., shorter dwell times) across the entire conformational landscape. 2) It tightens the distributions of conformational states and dwell times. 3) It causes CLeNOSr to favor more open conformational states, and to populate a distinct more open conformational state, which may correlate with the output state during catalysis of the NO synthesis reaction. These three CaM effects could increase electron flux through the reductase domain to both cytochrome c and the eNOS oxygenase domain heme.
Due to an absence of the heme-containing oxygenase domain in our truncated eNOSr protein, our single-molecule experiments exclude any possible influence from an interaction between the oxygenase domain and CaM, which could conceivably cause some differences in the activity or dynamics between the eNOSr and full-length eNOS proteins. Nevertheless, our single-molecule experimental results are still well correlated with recent structural studies of full-length NOS proteins (4,5,14,23,24) in their indicating that CaM binding generates an extra new open state of eNOSr, which we believe relates to the state in which the FMN subdomain undergoes a large rotation and becomes available for oxygenase domain interactions as observed in the recent CryoEM studies. In both the eNOSr and full-length proteins, CaM binding may induce similar flexibility that allows its FMN subdomain to be released and undergo a large rotation that enables electron transfer to the oxygenase domain heme.
To conclude, obtaining coordinate information on conformational states and dwell times in a multidomain redox enzyme, at a single-molecule level, is a powerful means to define the enzyme’s conformational behavior and to investigate how its conformational behavior relates to catalysis, and in the case of NOS enzymes how these facets are regulated by external players such as CaM. Based on our 3D plots of the single-molecule FRET time trajectories, we observed CLeNOSr to predominantly populate two conformational states representing an intermediate open and a closed state, and with CaM binding to populate an additional more open conformational state critical for electron transfer in the overall enzymatic activity. Our observing three conformational states with CaM binding presents the first direct evidence under real-time measurements for these functional conformational states, which had been reported in single-particle EM measurements of the 3D structure of mammalian NOSs. These conformational states and associated shorter dwell times induced by CaM binding are likely important for electron transfer functions and NO synthesis, and we now are capable of probing their populations, and fluctuation dynamics. Our current results shed new light on how CaM and eNOS function in biological signaling cascades by revealing how their interaction directs the conformational behavior of eNOS, which in turn enables the electron transfer reactions that are required to support NO synthesis.
Author contributions
H.P.L. and D.J.S. designed the research and contributed the analytical tools. Y.H. and M.M.H. performed research. Y.H., M.M.H., D.J.S., and H.P.L. analyzed data and wrote the article.
Acknowledgments
The study was supported by National Institutes of Health Grants GM51491 (D.J.S. and H.P.L.) and HL076491 (D.J.S.) and support from the Ohio Eminent Scholar Endowment (H.P.L.).
Editor: Alexandr Kornev.
Footnotes
Mohammad. M. Haque’s present address is Department of Biotechnology, Jamia Millia Islamia University, New Delhi, India
Yufan He and Mohammad Mahfuzul Haque contributed equally to this work.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.11.001.
Contributor Information
Dennis J. Stuehr, Email: stuehrd@ccf.org.
H. Peter Lu, Email: hplu@bgsu.edu.
Supporting material
Proposed model for CaM-mediated regulation of eNOS. In the absence of CaM, the enzyme mainly populates two conformational states: a closed input state with a broad range of dwell times in which the FMN domain is in contact with the NADPH/FAD domain so it can receive electrons, and an intermediate open state that has relatively shorter dwell times where the FMN domain is free from the NADPH/FAD domain. With the binding of CaM, the enzyme populates a third more open output conformational state, and the dwell times in all three conformational states are considerably shortened. In CaM-bound eNOS, the three conformational states likely represent the FMN domain electron input state, an intermediate open state, and a more open FMN domain electron output state in which the FMN domain is in contact with the heme-containing eNOSoxy domain. The intermediate conformational state is populated during the transitions between the input and output states.
References
- 1.Daff S. NO synthase: structures and mechanisms. Nitric Oxide. 2010;23:1–11. doi: 10.1016/j.niox.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Poulos T.L. Heme enzyme structure and function. Chem. Rev. 2014;114:3919–3962. doi: 10.1021/cr400415k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stuehr D.J., Tejero J., Haque M.M. Structural and mechanistic aspects of flavoproteins: electron transfer through the nitric oxide synthase flavoprotein domain. FEBS J. 2009;276:3959–3974. doi: 10.1111/j.1742-4658.2009.07120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell M.G., Smith B.C., et al. Marletta M.A. Molecular architecture of mammalian nitric oxide synthases. Proc. Natl. Acad. Sci. USA. 2014;111:E3614–E3623. doi: 10.1073/pnas.1413763111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith B.C., Underbakke E.S., et al. Marletta M.A. Nitric oxide synthase domain interfaces regulate electron transfer and calmodulin activation. Proc. Natl. Acad. Sci. USA. 2013;110:E3577–E3586. doi: 10.1073/pnas.1313331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sobolewska-Stawiarz A., Leferink N.G.H., et al. Scrutton N.S. Energy landscapes and catalysis in nitric-oxide synthase. J. Biol. Chem. 2014;289:11725–11738. doi: 10.1074/jbc.M114.548834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcin E.D., Bruns C.M., et al. Getzoff E.D. Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase. J. Biol. Chem. 2004;279:37918–37927. doi: 10.1074/jbc.M406204200. [DOI] [PubMed] [Google Scholar]
- 8.Ghosh D.K., Salerno J.C. Nitric oxide synthases: domain structure and alignment in enzyme function and control. Front. Biosci. 2003;8:d193–d209. doi: 10.2741/959. [DOI] [PubMed] [Google Scholar]
- 9.Daff S. Calmodulin-dependent regulation of mammalian nitric oxide synthase. Biochem. Soc. Trans. 2003;31:502–505. doi: 10.1042/bst0310502. [DOI] [PubMed] [Google Scholar]
- 10.Dunford A.J., Rigby S.E.J., et al. Scrutton N.S. Conformational and thermodynamic control of electron transfer in neuronal nitric oxide synthase. Biochemistry. 2007;46:5018–5029. doi: 10.1021/bi7001339. [DOI] [PubMed] [Google Scholar]
- 11.Feng C., Tollin G., et al. Ghosh D.K. Intraprotein electron transfer in a two-domain construct of neuronal nitric oxide synthase: the output state in nitric oxide formation. Biochemistry. 2006;45:6354–6362. doi: 10.1021/bi060223n. [DOI] [PubMed] [Google Scholar]
- 12.Guan Z.W., Iyanagi T. Electron transfer is activated by calmodulin in the flavin domain of human neuronal nitric oxide synthase. Arch. Biochem. Biophys. 2003;412:65–76. doi: 10.1016/s0003-9861(03)00009-2. [DOI] [PubMed] [Google Scholar]
- 13.Ilagan R.P., Tiso M., et al. Stuehr D.J. Differences in a conformational equilibrium distinguish catalysis by the endothelial and neuronal nitric-oxide synthase flavoproteins. J. Biol. Chem. 2008;283:19603–19615. doi: 10.1074/jbc.M802914200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Persechini A., Tran Q.K., et al. Gogol E.P. Calmodulin-induced structural changes in endothelial nitric oxide synthase. FEBS Lett. 2013;587:297–301. doi: 10.1016/j.febslet.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salerno J.C., Ray K., et al. Ghosh D.K. Calmodulin activates neuronal nitric oxide synthase by enabling transitions between conformational states. FEBS Lett. 2013;587:44–47. doi: 10.1016/j.febslet.2012.10.039. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh D.K., Ray K., et al. Salerno J.C. FMN fluorescence in inducible NOS constructs reveals a series of conformational states involved in the reductase catalytic cycle. FEBS J. 2012;279:1306–1317. doi: 10.1111/j.1742-4658.2012.08525.x. [DOI] [PubMed] [Google Scholar]
- 17.Pudney C.R., Khara B., et al. Scrutton N.S. Coupled motions direct electrons along human microsomal P450 Chains. PLoS Biol. 2011;9:e1001222. doi: 10.1371/journal.pbio.1001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Craig D.H., Chapman S.K., Daff S. Calmodulin activates electron transfer through neuronal nitric-oxide synthase reductase domain by releasing an NADPH-dependent conformational lock. J. Biol. Chem. 2002;277:33987–33994. doi: 10.1074/jbc.M203118200. [DOI] [PubMed] [Google Scholar]
- 19.Gachhui R., Presta A., et al. Stuehr D.J. Characterization of the reductase domain of rat neuronal nitric oxide synthase generated in the methylotrophic yeast Pichia pastoris. Calmodulin response is complete within the reductase domain itself. J. Biol. Chem. 1996;271:20594–20602. doi: 10.1074/jbc.271.34.20594. [DOI] [PubMed] [Google Scholar]
- 20.Huang W.C., Ellis J., et al. Roberts G.C.K. Redox-linked domain movements in the catalytic cycle of cytochrome p450 reductase. Structure. 2013;21:1581–1589. doi: 10.1016/j.str.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnett D.C., Persechini A., et al. Johnson C.K. Fluorescence quenching studies of structure and dynamics in calmodulin-eNOS complexes. FEBS Lett. 2015;589:1173–1178. doi: 10.1016/j.febslet.2015.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollingsworth S.A., Holden J.K., et al. Poulos T.L. Elucidating nitric oxide synthase domain interactions by molecular dynamics. Protein sci. 2016;25:374–382. doi: 10.1002/pro.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volkmann N., Martásek P., et al. Masters B.S. Holoenzyme structures of endothelial nitric oxide synthase - an allosteric role for calmodulin in pivoting the FMN domain for electron transfer. J. Struct. Biol. 2014;188:46–54. doi: 10.1016/j.jsb.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yokom A.L., Morishima Y., et al. Southworth D.R. Architecture of the nitric-oxide synthase holoenzyme reveals large conformational changes and a calmodulin-driven release of the FMN domain. J. Biol. Chem. 2014;289:16855–16865. doi: 10.1074/jbc.M114.564005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ilagan R.P., Tejero J., et al. Stuehr D.J. Regulation of FMN subdomain interactions and function in neuronal nitric oxide synthase. Biochemistry. 2009;48:3864–3876. doi: 10.1021/bi8021087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konas D.W., Zhu K., et al. Stuehr D.J. The FAD-shielding residue Phe1395 regulates neuronal nitric-oxide synthase catalysis by controlling NADP+ affinity and a conformational equilibrium within the flavoprotein domain. J. Biol. Chem. 2004;279:35412–35425. doi: 10.1074/jbc.M400872200. [DOI] [PubMed] [Google Scholar]
- 27.Feng C., Tollin G., et al. Ghosh D.K. Direct measurement by laser flash photolysis of intraprotein electron transfer in a rat neuronal nitric oxide synthase. J. Am. Chem. Soc. 2007;129:5621–5629. doi: 10.1021/ja068685b. [DOI] [PubMed] [Google Scholar]
- 28.He Y., Haque M.M., et al. Lu H.P. Single-molecule spectroscopy reveals how calmodulin activates NO synthase by controlling its conformational fluctuation dynamics. Proc. Natl. Acad. Sci. USA. 2015;112:11835–11840. doi: 10.1073/pnas.1508829112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stuehr D.J., Santolini J., et al. Adak S. Update on mechanism and catalytic regulation in the NO synthases. J. Biol. Chem. 2004;279:36167–36170. doi: 10.1074/jbc.R400017200. [DOI] [PubMed] [Google Scholar]
- 30.Haque M.M., Panda K., et al. Stuehr D.J. A connecting hinge represses the activity of endothelial nitric oxide synthase. Proc. Natl. Acad. Sci. USA. 2007;104:9254–9259. doi: 10.1073/pnas.0700332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roman L.J., Masters B.S.S. Electron transfer by neuronal nitric-oxide synthase is regulated by concerted interaction of calmodulin and two intrinsic regulatory elements. J. Biol. Chem. 2006;281:23111–23118. doi: 10.1074/jbc.M603671200. [DOI] [PubMed] [Google Scholar]
- 32.Haque M.M., Fadlalla M.A., et al. Stuehr D.J. Control of electron transfer and catalysis in neuronal nitric-oxide synthase (nNOS) by a hinge connecting its FMN and FAD-NADPH domains. J. Biol. Chem. 2012;287:30105–30116. doi: 10.1074/jbc.M112.339697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haque M.M., Kenney C., et al. Stuehr D.J. A kinetic model linking protein conformational motions, interflavin electron transfer and electron flux through a dual-flavin enzyme-simulating the reductase activity of the endothelial and neuronal nitric oxide synthase flavoprotein domains. FEBS J. 2011;278:4055–4069. doi: 10.1111/j.1742-4658.2011.08310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sucharitakul J., Chaiyen P., et al. Ballou D.P. Kinetic mechanisms of the oxygenase from a two-component enzyme, p-hydroxyphenylacetate 3-hydroxylase from Acinetobacter baumannii. J. Biol. Chem. 2006;281:17044–17053. doi: 10.1074/jbc.M512385200. [DOI] [PubMed] [Google Scholar]
- 35.Roy R., Hohng S., Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selvin P.R., Ha T. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2008. Single-Molecule Techniques: A Laboratory Manual. [Google Scholar]
- 37.Chen Y., Hu D.H., et al. Lu H.P. Probing single-molecule T4 lysozyme conformational dynamics by intramolecular fluorescence energy transfer. J. Phys. Chem. B. 2003;107:7947–7956. [Google Scholar]
- 38.He Y., Li Y., et al. Lu H.P. Probing single-molecule enzyme active-site conformational state intermittent coherence. J. Am. Chem. Soc. 2011;133:14389–14395. doi: 10.1021/ja204644y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu H.P., Xun L., Xie X.S. Single-molecule enzymatic dynamics. Science. 1998;282:1877–1882. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 40.Liu R., Hu D., et al. Lu H.P. Revealing two-state protein-protein interactions of calmodulin by single-molecule spectroscopy. J. Am. Chem. Soc. 2006;128:10034–10042. doi: 10.1021/ja057005m. [DOI] [PubMed] [Google Scholar]
- 41.He Y., Lu M., Lu H.P. Single-molecule photon stamping FRET spectroscopy study of enzymatic conformational dynamics. Phys. Chem. Chem. Phys. 2013;15:770–775. doi: 10.1039/c2cp42944f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haque M.M., Bayachou M., et al. Stuehr D.J. Distinct conformational behaviors of four mammalian dual-flavin reductases (cytochrome P450 reductase, methionine synthase reductase, neuronal nitric oxide synthase, endothelial nitric oxide synthase) determine their unique catalytic profiles. FEBS J. 2014;281:5325–5340. doi: 10.1111/febs.13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamdane D., Xia C., et al. Waskell L. Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450. J. Biol. Chem. 2009;284:11374–11384. doi: 10.1074/jbc.M807868200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haque M.M., Ray S.S., Stuehr D.J. Phosphorylation controls endothelial nitric-oxide synthase by regulating its conformational dynamics. J. Biol. Chem. 2016;291:23047–23057. doi: 10.1074/jbc.M116.737361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haque M.M., Bayachou M., et al. Stuehr D.J. Charge-pairing interactions control the conformational setpoint and motions of the FMN domain in neuronal nitric oxide synthase. Biochem. J. 2013;450:607–617. doi: 10.1042/BJ20121488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia C., Misra I., et al. Kim J.J. Regulation of interdomain interactions by calmodulin in inducible nitric-oxide synthase. J. Biol. Chem. 2009;284:30708–30717. doi: 10.1074/jbc.M109.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Astashkin A.V., Feng C. Solving kinetic equations for the laser flash photolysis experiment on nitric oxide synthases: effect of conformational dynamics on the interdomain electron transfer. J. Phys. Chem. A. 2015;119:11066–11075. doi: 10.1021/acs.jpca.5b08414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng C., Chen L., et al. Sun X. Dissecting regulation mechanism of the FMN to heme interdomain electron transfer in nitric oxide synthases. J. Inorg. Biochem. 2014;130:130–140. doi: 10.1016/j.jinorgbio.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng C. Mechanism of nitric oxide synthase regulation: electron transfer and interdomain interactions. Coord. Chem. Rev. 2012;256:393–411. doi: 10.1016/j.ccr.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proposed model for CaM-mediated regulation of eNOS. In the absence of CaM, the enzyme mainly populates two conformational states: a closed input state with a broad range of dwell times in which the FMN domain is in contact with the NADPH/FAD domain so it can receive electrons, and an intermediate open state that has relatively shorter dwell times where the FMN domain is free from the NADPH/FAD domain. With the binding of CaM, the enzyme populates a third more open output conformational state, and the dwell times in all three conformational states are considerably shortened. In CaM-bound eNOS, the three conformational states likely represent the FMN domain electron input state, an intermediate open state, and a more open FMN domain electron output state in which the FMN domain is in contact with the heme-containing eNOSoxy domain. The intermediate conformational state is populated during the transitions between the input and output states.