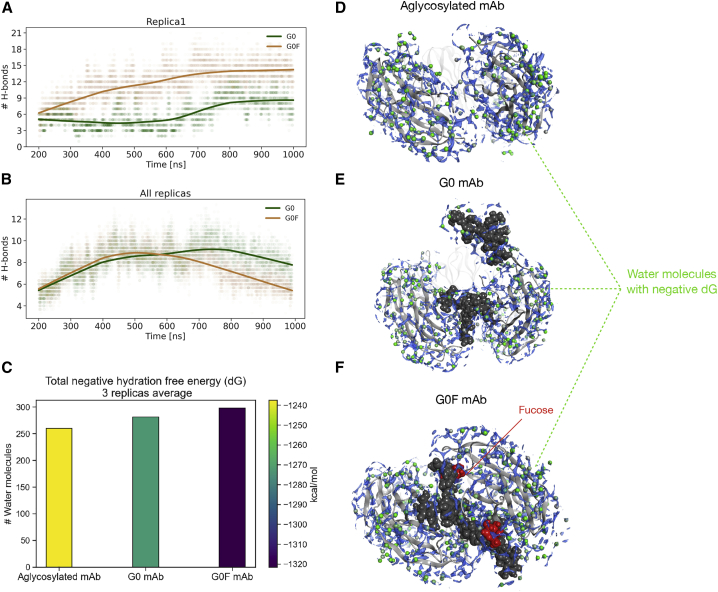
Figure 7.
H-bond number and solvent analysis. (A) The H-bond number between G0 or G0F chains (A and B) and the antibody versus simulation time. The first 200 ns were excluded from calculation. (B) The mean H-bond number between G0 or G0F chains (A and B) and the antibody computed for replicas. For both the scatter plots, a lowess nonparametric interpolation was used. (C–F) Solvent analysis performed by 3D-RISM on aglycosylated, G0, and G0F mAbs. In (C), a bar chart showing the mean number of predicted water molecules associated with a negative dG and the average negative dG of three replicas is given. Bars are colored according to the energy colormap. In (D)–(F), the top view of CH2 domains is shown as gray ribbons and glycans as dark gray spheres, with fucose in red. The dG isolevel density is shown in blue, and water molecules with a negative dG value are shown as green spheres. To see this figure in color, go online.