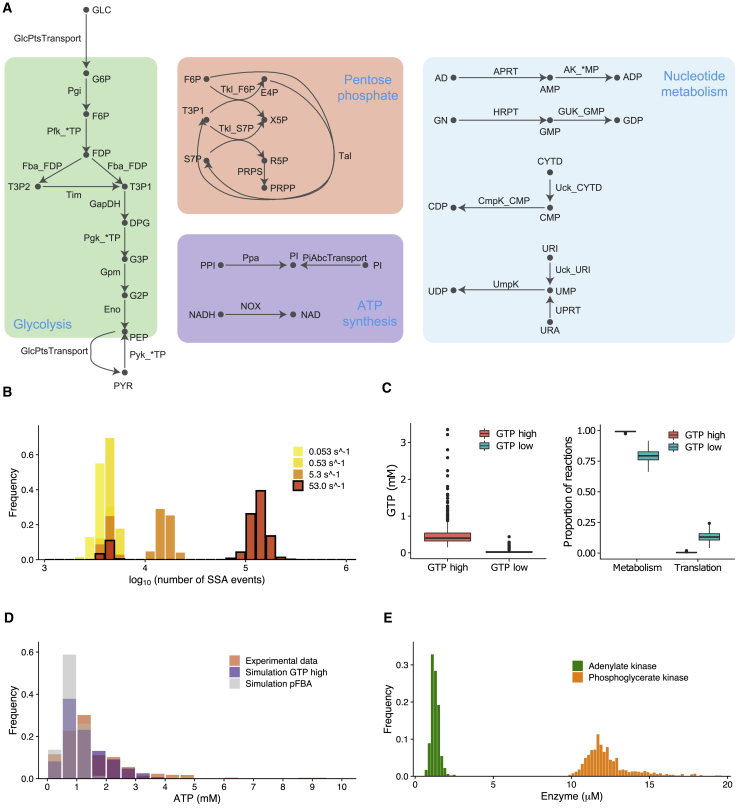
Figure 4.
Single-cell simulation of M. pneumoniae metabolism using SSA-FBA. (A) Metabolic reaction network used in the model annotated with species listed in Supporting materials and methods. (B) Comparison of distributions of the number of SSA execution events per simulation for different values of the GMK reaction rate constant. For reference, the bimodal distribution corresponding to kcat = 53.0 s−1 is outlined in black. (C) Box plots showing significant differences between GTP concentrations (p < 2.2 × 10−16) and the relative proportions of reaction execution events that correspond to metabolic reactions (p < 2.2 × 10−16) and translation reactions (p < 2.2 × 10−16), respectively, between the low and high GTP groups. (D) Distributions of time-averaged ATP concentrations from simulations with the GTP-high group and pFBA compared with the experimental distribution from (52). (E) Distribution of time-averaged ADK and PGK concentrations from simulations with the GTP-high group. To see this figure in color, go online.