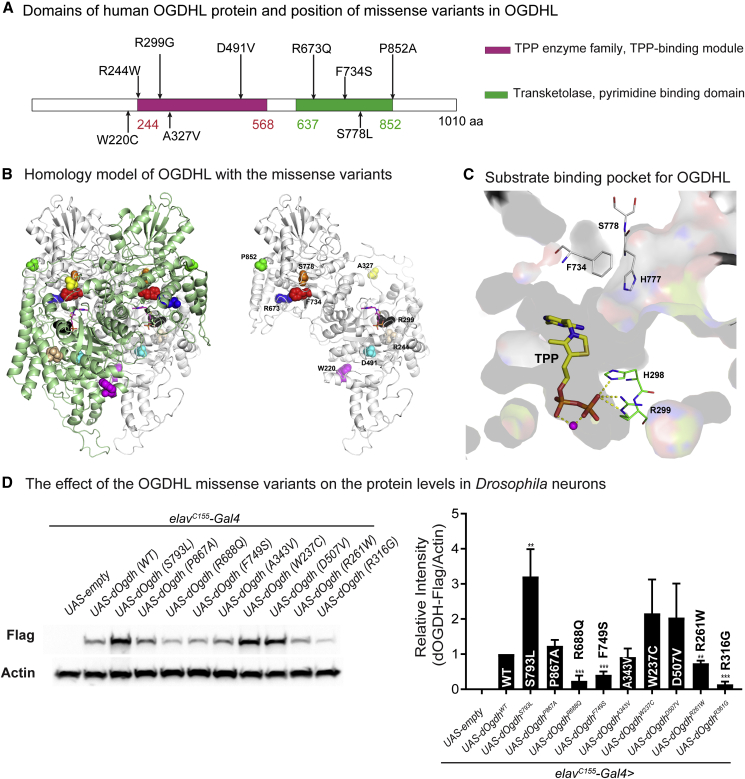
Figure 3.
Protein modeling of OGDHL dimer and the effects of eight missense variants on protein levels
(A) Schematic representation of protein domains of human OGDHL and positions of the eight missense variants together with the previously identified p.Ser778Leu variant. Pink indicates a TPP-binding domain. Green indicates a transketolase domain.
(B) In silico protein structure prediction of human OGDHL dimer and position of mutated residues. Dimer of OGDHL is shown in ribbon (left). Each monomer is colored with green and white. Bound TPP is shown in stick model and mutation sites are in CPK model. One monomer is shown for clarity (right).
(C) Only thin section of substrate binding pocket was shown with surface rendering for clarity. Amino acid residues involved in missense variants in the pocket are shown in stick model together with TPP. Residues from cis subunit are shown in green and residues from trans in gray. Bound magnesium is shown in magenta ball.
(D) Immunoblots for Drosophila heads expressing wild-type dOgdh-FLAG or dOgdh-FLAG carrying the homologous missense variants identified from individuals with bi-allelic OGDHL variants. Three biological replicates were quantified. Error bars represent SEM. p values were obtained by ANOVA. ∗∗p < 0.01, ∗∗∗p < 0.001.