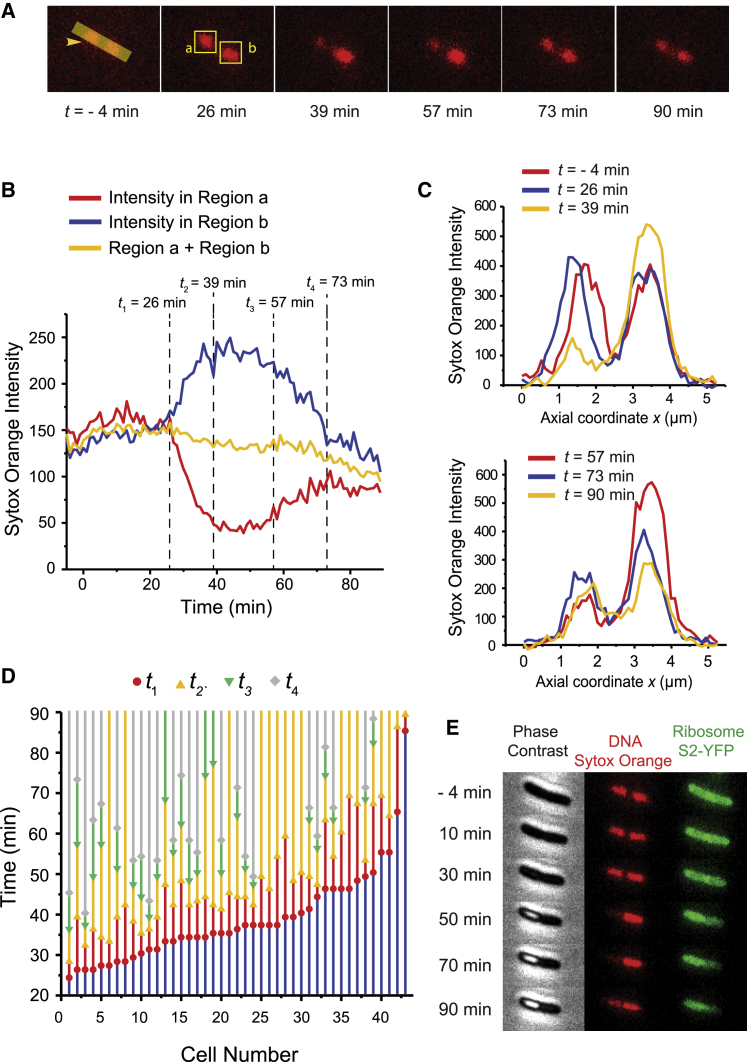
Figure 3.
Effects of MM-CH on nucleoid morphology at long times. Chromosomal DNA was stained with 500 nM Sytox Orange for 10 min before imaging. Flow of 2× MIC MM-CH began at t = 0. Images acquired at 1 min per frame. (A) Sytox Orange fluorescence snapshots of single E. coli cell at different times. (B) Sytox Orange intensity versus time for the same cell shown in (A). Total Sytox Orange intensity from the entire cell (yellow curve), from the left nucleoid lobe (red curve, region a as in A), and from the right nucleoid lobe (blue curve, region b as in A). Total intensity from the entire cell is scaled by a factor of 1.6. Labeled times are t1 (beginning of coalescence of nucleoid lobes), t2 (beginning of phase 2, the nucleoid reaches its most compact state), t3 (beginning of phase 3, partial recovery), and t4 (nucleoids have reached the apparent final state). (C) Projected axial Sytox Orange intensity profile along the cell long axis (arrowhead in A) at different times. Left nucleoid lobe merged to the other one and partially recovered at later times. (D) The timeline of events t1–t4 for 43 cells from four experiments. (E) Phase contrast, Sytox Orange fluorescence and ribosome S2-YFP snapshots of single E. coli cell at different times after flowing MM-CH.