Abstract
Background & Aims
Inactivating mutations of KDM6A, a histone demethylase, were frequently found in pancreatic ductal adenocarcinoma (PDAC). We investigated the role of KDM6A (lysine demethylase 6A) in PDAC development.
Methods
We performed a pancreatic tissue microarray analysis of KDM6A protein levels. We used human PDAC cell lines for KDM6A knockout and knockdown experiments. We performed bromouridine sequencing analysis to elucidate the effects of KDM6A loss on global transcription. We performed studies with Ptf1aCre; LSL-KrasG12D; Trp53R172H/+; Kdm6afl/fl or fl/Y, Ptf1aCre; Kdm6afl/fl or fl/Y, and orthotopic xenograft mice to investigate the impacts of Kdm6a deficiency on pancreatic tumorigenesis and pancreatitis.
Results
Loss of KDM6A was associated with metastasis in PDAC patients. Bromouridine sequencing analysis showed up-regulation of the epithelial–mesenchymal transition pathway in PDAC cells deficient in KDM6A. Loss of KDM6A promoted mesenchymal morphology, migration, and invasion in PDAC cells in vitro. Mechanistically, activin A and subsequent p38 activation likely mediated the role of KDM6A loss. Inhibiting either activin A or p38 reversed the effect. Pancreas-specific Kdm6a-knockout mice pancreata showed accelerated PDAC progression, developed a more aggressive undifferentiated type of PDAC, and increased metastases in the background of Kras and p53 mutations. Kdm6a-deficient pancreata in a pancreatitis model had a delayed recovery with increased PDAC precursor lesions compared with wild-type pancreata.
Conclusions
Loss of KDM6A accelerates PDAC progression and metastasis, most likely by a noncanonical p38-dependent activin A pathway. KDM6A also promotes pancreatic tissue recovery from pancreatitis. Activin A might be used as a therapeutic target for KDM6A-deficient PDACs.
Keywords: Epigenetics, Cancer Cell Plasticity, Metastasis, Activin A
Abbreviations used in this paper: ADM, acinar-to-ductal metaplasia; Bru-seq, bromouridine sequencing; ELISA, enzyme-linked immunosorbent assay; EMT, epithelial-mesenchymal transition; H3K27, histone 3 lysine 27; IHC, immunohistochemistry; INHBA, Inhibin Subunit Beta A; KC, Ptf1aCre;LSL-KrasG12D/+; KO, knockout; KPC, Ptf1aCre;LSL-KrasG12D;LSL-p53R172H; KRAS, Kirsten rat sarcoma virus; PanIN, pancreatic intraepithelial neoplasia; PDAC, pancreatic ductal adenocarcinoma; TCGA, the Cancer Genome Atlas; TGF-β, transforming growth factor-β; TMA, tissue microarray
Graphical abstract
Summary.
Loss of KDM6A promotes pancreatic cancer progression and metastasis and delays pancreatic tissue recovery from pancreatitis. Mechanistically, a noncanonical p38-dependent activin A pathway likely mediates KDM6A function.
Pancreatic ductal adenocarcinoma (PDAC) is a lethal disease and is predicted to become the second leading cause of cancer death in the United States by 2030, with an overall 5-year survival rate of less than 10%.1 One of the main reasons for the dismal survival rate is early metastasis.2 Studies have found limited heterogeneity of driver gene mutations in PDAC metastatic founder subclones.3, 4, 5 These founder subclones also are present in the primary tumor, suggesting that factors other than genetic mutations may contribute to metastasis. Recent studies have suggested that aberrant epigenetic regulation may be one of the crucial mechanisms for tumor progression and early metastasis.6 Although PDAC is characterized by common mutations such as KRAS, TP53, CDKN2A, and SMAD4, approximately a quarter of the PDACs contain mutations in epigenetic and chromatin remodeling genes.7,8 Some of the most frequently mutated epigenetic genes, such as KDM6A and KMT2D, encode histone modification enzymes. However, how exactly the alterations of these genes impact PDAC progression is not entirely known.
Lysine demethylase 6A (KDM6A), also known as Ubiquitously-Transcribed X Chromosome Tetratricopeptide Repeat Protein (UTX), is a histone 3 lysine 27 (H3K27) demethylase that regulates gene transcription. Recent whole-genome sequencing analysis of PDAC showed frequent mutations in KDM6A.7,8 Inactivating mutations of KDM6A were found in up to 18% of PDACs, suggesting a tumor-suppressive role of KDM6A.8 KDM6A mutations have been observed across a broad range of cancers.9 In myeloid leukemia, bladder cancer, and breast cancer, KDM6A was found to play a tumor-suppressive role10, 11, 12; while a different pro-oncogenic function has been described in cervical cancer.13 In breast cancer, KDM6A deficiency activates the transcription factor GATA3 and promotes cancer development and metastasis, possibly via activating the epithelial–mesenchymal transition (EMT) in vitro.14 Recent findings showed that loss of KDM6A induces gender-specific, squamous-like PDAC in genetic mouse models,15 and characterizes a poor prognostic subtype of human PDAC.16 However, the specific functions of KDM6A in PDAC development and metastasis, as well as the underlying mechanisms, have not been fully elucidated.
In this study, we sought to define the underlying mechanisms by which KDM6A loss contributes to PDAC development using both in vitro and in vivo experimental models. We unveiled a novel link between KDM6A and noncanonical activin A signaling as a major mechanism of cancer cell plasticity, tumorigenesis, and progression. These findings will pave the way for the discovery of new therapeutic targets in tumors with KDM6A inactivating mutations.
Results
Decreased KDM6A Correlates With Worse Prognosis in PDAC Patients
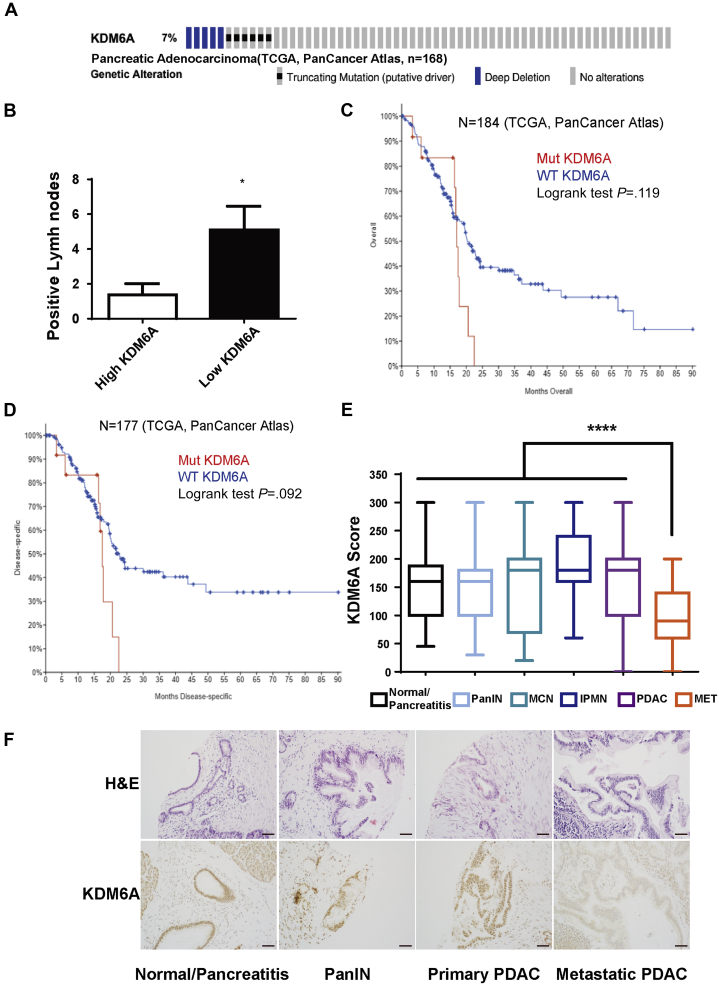
To investigate the frequency of genetic alterations of KDM6A in human PDAC, we queried The Cancer Genome Atlas (TCGA) (n = 168) and found that 7% of human PDACs carried truncating mutations and deep deletions of KDM6A (Figure 1A). In addition, patients with KDM6A-low PDACs had significantly higher numbers of lymph node metastasis compared with KDM6A-high PDACs (Figure 1B). Furthermore, patients with mutated KDM6A tended to have shorter overall and disease-specific survival compared with those with wild-type KDM6A (Figure 1C and D), albeit this was not statistically significant owing to a low sample number. To determine whether KDM6A protein expression also is decreased in human primary and metastatic PDACs, we used a tissue microarray (TMA) containing 213 duplicated human pancreatic tissue cores of normal/pancreatitis (n = 36), precursor lesions (pancreatic intraepithelial neoplasia [PanIN], intraductal papillary mucinous neoplasm, mucinous cystic neoplasm) (n = 84), and primary and metastatic PDACs (n = 93) (Table 1). Immunohistochemistry (IHC) staining showed that KDM6A expression was significantly lower in metastatic PDAC compared with normal/pancreatitis and all other lesions, including primary PDAC (P < .0001) (Figure 1E and F, Table 1). Collectively, these results showed that loss of KDM6A was associated with worse prognosis and metastasis in human PDAC.
Figure 1.
KDM6A mutations and expression profiles and their association with survival in human PDACs. (A) Sequencing data of human PDACs (N = 168) were queried from TCGA PanCancer Atlas database and PDACs with KDM6A mutations are highlighted by various colored bars. (B) Number of lymph nodes positive for metastatic PDAC in patients with high- or low-KDM6A-expressing PDACs (TCGA, N = 22). (C and D) Kaplan–Meier survival curves of (C) overall and (D) disease-specific survival in patients with KDM6A-mutated (Mut) or wild-type (WT) PDAC based on TCGA PanCancer Atlas data sets. (E) IHC staining scores of KDM6A in normal/pancreatitis (n = 36), PanIN (n = 32), mucinous cystic neoplasm (MCN, n = 13), intraductal papillary mucinous neoplasm (IPMN, n = 39), primary PDACs (PDAC, n = 74) and metastatic PDACs (MET, n = 19) from a human pancreas TMA. (F) Representative images of H&E and KDM6A IHC staining of the pancreas TMA with pancreata in different stages of PDAC progression. ∗P < .05, ∗∗∗∗P < .0001, unpaired t test.
Table 1.
Protein Expression of KDM6A in Human PDAC and its Precursor Lesions
| Category | N | Means ± SD | P (MET vs others) |
|---|---|---|---|
| Normal/pancreatitis | 36 | 153 ± 57 | |
| PanIN | 32 | 155 ± 67 | |
| MCN | 13 | 159 ± 90 | |
| IPMN | 39 | 194 ± 65 | |
| PDAC | 74 | 166 ± 68 | |
| MET PDAC | 19 | 100 ± 53 | .00003 |
| Total | 213 |
IPMN, intraductal papillary mucinous neoplasm; MCN, mucinous cystic neoplasm; MET, metastatic.
KDM6A Modulates PDAC Cell Identity, Tumor Sphere Formation, Migration, and Invasion
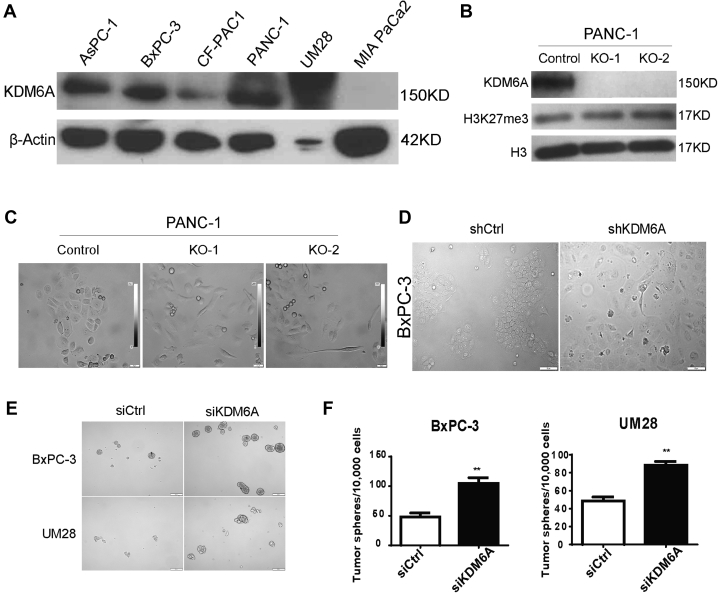
To investigate the biological functions of KDM6A in PDAC progression, we first surveyed the protein expression levels of KDM6A in several commonly used human PDAC cell lines. We were able to detect KDM6A in multiple PDAC cell lines, including AsPC-1, BxPC-3, CF-PAC1, PANC-1, and UM28 (Figure 2A). Of note, we observed especially abundant levels of KDM6A in PANC-1 and UM28, but not in MIA PaCa-2. We then knocked out KDM6A in PANC-1 cells using Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) technology (Figure 2B). Because KDM6A is a H3K27 demethylase, we assessed the total H3K27me3 levels in KDM6A-knockout (KO) cells. Interestingly, KDM6A loss did not significantly alter H3K27me3 levels (Figure 2B).
Figure 2.
KDM6A modulates PDAC cell identity and tumor sphere formation. (A) Western blot of KDM6A in 6 human PDAC cell lines. β-actin was used as loading control. (B) Western blot of KDM6A, H3K27me3, and H3 in control and 2 different clones of KDM6A-KO PANC-1 cells (KO-1 and KO-2). (C) Representative phase-contrast microscopic images of control and KDM6A-KO PANC-1 cells. (D) Representative microscopic phase-contrast images of control or KDM6A short hairpin (sh) RNA knockdown BxPC-3 cells. Scale bar: 50 μm. (E) Representative images and (F) quantification of tumor spheres formed in BxPC-3 and UM28 cells transfected with control or KDM6A small interfering (si) RNAs. ∗∗P < .01, unpaired t test. Ctrl, control.
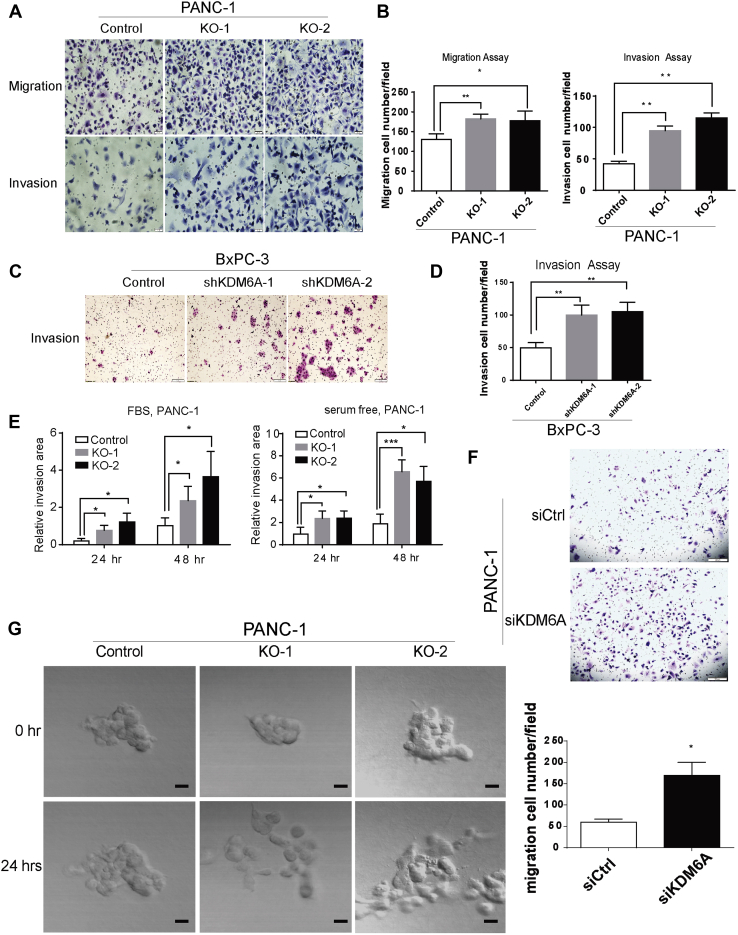
We then sought to determine the biologic impact of KDM6A loss in PDAC cells. We observed a cell morphology change from epithelial (polygonal-shaped and cohesive) to mesenchymal (spindled-shaped and discohesive) phenotype in both KDM6A-KO and knockdown PDAC cell lines (Figure 2C and D). Knockdown of KDM6A also promoted tumor sphere formation (Figure 2E and F). We then used the Transwell assay and a 3D culture system with live microscopy monitoring to determine if KDM6A loss affects cell migration and invasion. KDM6A loss significantly promoted cell migration and invasion in both models (Figure 3). Taken together, these findings implied that loss of KDM6A promoted the mesenchymal identity, tumor sphere formation, cell migration, and invasion ability in PDAC cells.
Figure 3.
KDM6A loss promotes tumor cell migration and invasion. (A) Representative images and (B) quantification of cell migration and invasion assays in control and KDM6A-KO PANC-1 cells. (C) Representative images and (D) quantification of cell invasion assay in BxPC-3 cells transfected with control or 2 different KDM6A short hairpin (sh) RNAs. (E) Quantifications of cell invasion using 3-dimensional cell culture of control and KDM6A-KO PANC-1 cells growing in media either with or without fetal bovine serum (FBS) at 24 and 48 hours. (F) Representative images (top) and quantifications (bottom) of migrated control and KDM6A small interfering (si) RNA knockdown PANC-1 cells. (G) Representative pictures of cell migration comparing KDM6A-KO PANC-1 cells with control PANC-1 cells using a 3D cell culture model at the 0- and 24-hour time points. Scale bar: 100 μm. Live videos spanning cell migration over a 24-hour window are provided in the supplementary material. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001, unpaired t test. Ctrl, control.
KDM6A Loss Up-Regulates EMT and Activin A Expression
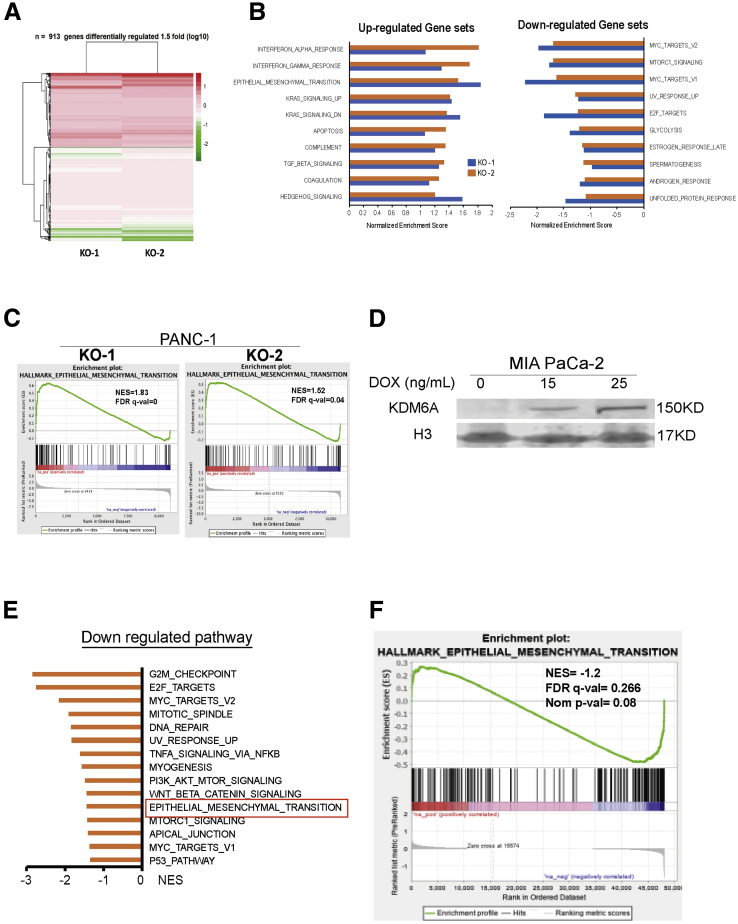
We then sought to understand the underlying mechanisms by which KDM6A loss promoted tumor. Using bromouridine sequencing (Bru-seq) technology,17 we measured the global nascent RNA synthesis alterations caused by loss of KDM6A. We chose Bru-seq because it measures newly synthesized RNA, not steady-state RNA. Therefore, Bru-seq catches a different pool of RNAs that reflects variable rates of active transcription and RNA degradation. A total of 913 genes were differentially regulated (>1.5-fold change compared with wild-type cells) in 2 KDM6A-KO clones (Figure 4A). Gene Set Enrichment Analysis showed that the EMT and transforming growth factor-β (TGF-β) pathways were 2 of the top up-regulated pathways in both KDM6A-KO clones (Figure 4B and C). Other up-regulated pathways included interferon response, Kirsten rat sarcoma virus (KRAS) signaling, apoptosis, and hedgehog pathways. On the other hand, myelocytomatosis oncogene product (MYC), mechanistic target of rapamycin (mTOR) complex 1 (MTORC1) signaling, ultraviolet (UV) response, E2 factor (E2F), glycolysis, and estrogen and androgen response pathways were among the most down-regulated pathways in the KDM6A-KO cells (Figure 4B). We also performed Bru-seq analysis in MIA PaCa-2 cells with the doxycycline-inducible KDM6A overexpression system (Figures 2A and 4D). Bru-seq and gene enrichment analyses showed that EMT was one of the most down-regulated pathways when KDM6A expression is rescued (Figure 4E and F).
Figure 4.
KDM6A deficiency up-regulates the EMT pathway by Bru-seq analysis. (A) Heat map of differentially regulated genes in 2 KDM6A-KO PANC-1 cell clones (KO-1 and KO-2) compared with control cells based on Bru-seq data. (B) Top 10 up-regulated and down-regulated hallmark pathways in KDM6A-KO PANC-1 cells compared with control cells identified by the Gene Set Enrichment Analysis of Bru-seq data. (C) EMT pathway signature enrichment plots in 2 KDM6A-KO PANC-1 clones. (D) Western blot of KDM6A in a Tet-on doxycycline (DOX)-inducible KDM6A overexpression MIA PaCa-2 cell line. Histone 3 (H3) was used as loading control. (E) The top down-regulated hallmark gene pathways in KDM6A re-expressed MIA PaCa-2 cells compared with control cells identified by the Gene Set Enrichment Analysis of Bru-seq data. (F) EMT pathway signature enrichment plot in KDM6A re-expressed MIA PaCa-2 cells. FDR q val, false-discovery rate; NES, normalized enrichment score; Nom, nominal.
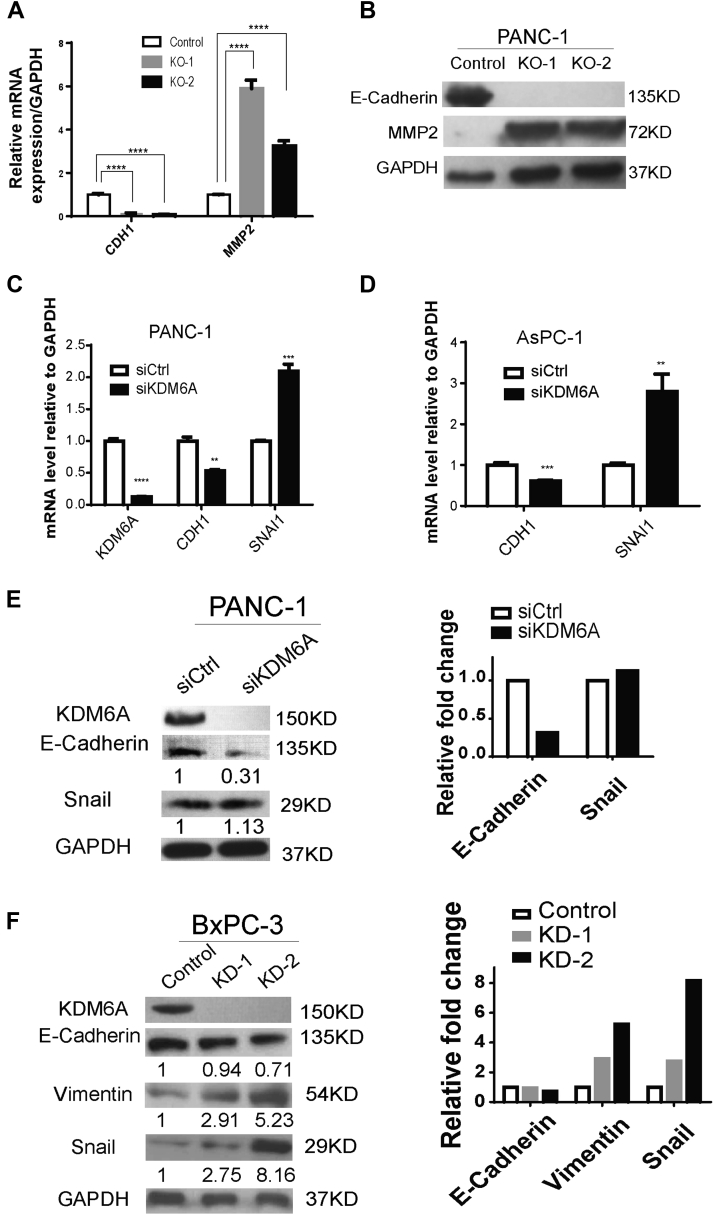
To confirm our Bru-seq data, we assessed the expression of CDH1 (encodes E-cadherin), a hallmark epithelial marker, and MMP2, a mesenchymal marker, in KDM6A-KO cells. Indeed, loss of KDM6A suppressed CDH1 expression while inducing MMP2 expression (Figure 5A and B). To determine if this observation applied to other PDAC cell lines and was not the result of off-target effects, we knocked down KDM6A in 3 different PDAC cell lines. Suppression of epithelial marker E-cadherin and up-regulation of mesenchymal markers Snail and/or vimentin were observed in KDM6A-knockdown cells compared with control cells in all these models (Figure 5C–F). These results support that loss of KDM6A induces the EMT process in PDAC cells.
Figure 5.
Confirmation of EMT up-regulation on KDM6A loss. (A) Quantitative real-time reverse-transcription polymerase chain reaction and (B) Western blot of CDH1 (E-cadherin) and MMP2 in control and KDM6A-KO PANC-1 cells. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as reference gene and loading control. (C) Real-time reverse-transcription polymerase chain reaction of CDH1 and SNAI1 in control and KDM6A small interfering (si) RNA knockdown PANC-1 and (D) AsPC-1 cells. GAPDH was used as a reference gene. (E) Western blot of KDM6A, E-cadherin, and Snail in control and KDM6A siRNA knockdown PANC-1 cells. Quantifications of E-cadherin and Snail immunoblot bands by ImageJ (National Institutes of Health, Bethesda, MD) were shown relative to siCtrl. Right: Relative fold changes of E-cadherin and Snail immunoblot bands compared with siCtrl. (F) Western blot of KDM6A, E-cadherin, Vimentin, and Snail in control and KDM6A short hairpin RNA knockdown BxPC-3 cells. GAPDH was used as loading control. Quantifications of E-cadherin, Vimentin, and Snail immunoblot bands by ImageJ were shown relative to control. Right: Relative fold changes of E-cadherin, Vimentin, and Snail immunoblot bands compared with control. ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001, unpaired t test. Ctrl, control; mRNA, messenger RNA.
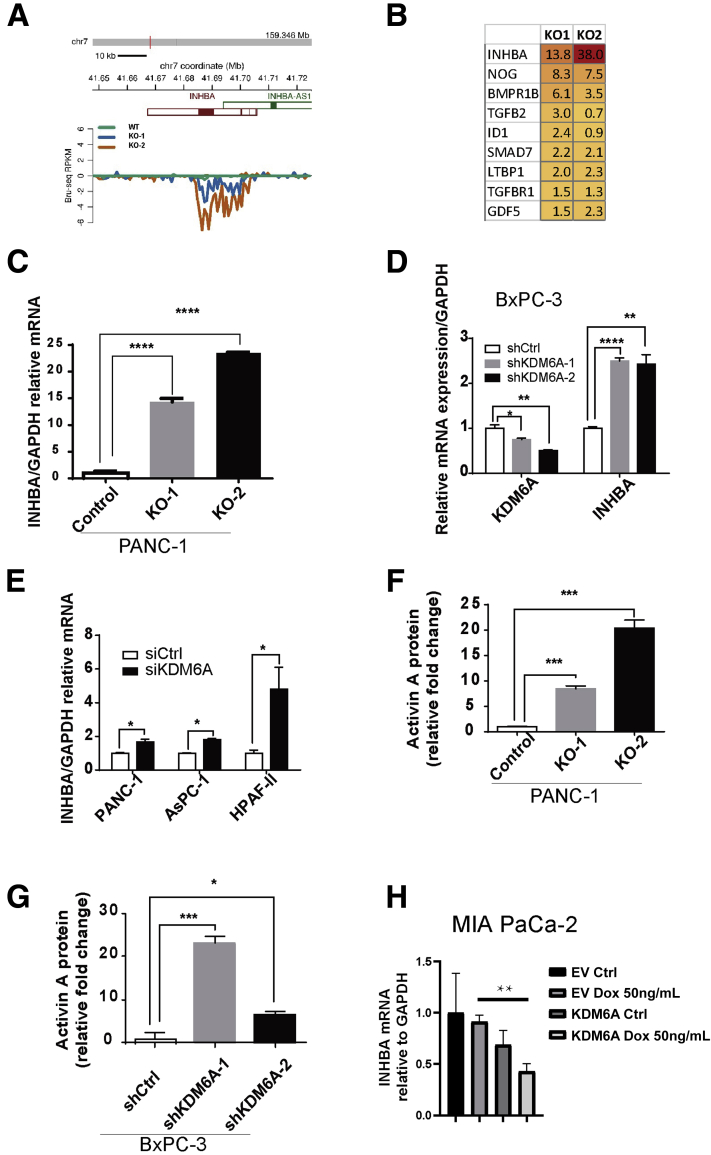
The most significantly up-regulated gene in the TGF-β pathway in both KDM6A-deficient clones was INHBA (Figure 6A and B). INHBA encodes activin A, a member of the TGF-β superfamily, which is well known to contribute to EMT, stemness, cell migration, and invasion.18,19 Therefore, we postulated that KDM6A signals through activin A. We first confirmed the up-regulation of activin A messenger RNA expression in both KDM6A-KO clones, which showed an approximately 14- to 23-fold increase (Figure 6C). To determine if this observation could be applied to other PDAC cell lines and was not caused by an off-target effect, we used short hairpin RNA or small interfering RNA to knockdown KDM6A in 4 different PDAC cell lines. INHBA was up-regulated in all 4 PDAC cell lines (Figure 6D and E). Because activin A is a secreted protein, we then used the enzyme-linked immunosorbent assay (ELISA) to measure activin A protein in the cell culture media. Either KO or knockdown of KDM6A led to a significant increase in the secreted activin A protein levels (Figure 6F and G). Finally, we assessed INHBA expression in our inducible KDM6A-overexpression cell line MIA PaCa-2. KDM6A overexpression led to decreased INHBA expression compared with control cells (Figure 6H). These results support that loss of KDM6A up-regulates activin A expression and secretion.
Figure 6.
KDM6A deficiency up-regulates activin A expression. (A) Reads per kilobase of transcript per million mapped reads (RPKM) of INHBA transcripts in KDM6A-KO and wild-type PANC-1 cells by Bru-seq analysis. (B) A heat map showing a list of genes in the TGF-β pathway that were up-regulated more than 1.5-fold in at least 1 clone of KDM6A-KO PANC-1 cells compared with control cells based on the Bru-seq data. (C) Real-time reverse-transcription polymerase chain reaction of INHBA in control and KDM6A-KO PANC-1 cells, (D) control and KDM6A short hairpin (sh) RNA knockdown BxPC-3 cells, and (E) control and KDM6A small interfering (si) RNA knockdown PANC-1, AsPC-1, and HPAF-II PDAC cell lines. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the reference gene. (F) Activin A protein levels in conditioned media from cultured control and KDM6A-KO PANC-1 cells and (G) shRNA control and KDM6A knockdown BxPC-3 cells measured by the enzyme-linked immunosorbent assay. (H) Real-time reverse-transcription polymerase chain reaction of INHBA in empty vector (EV) or a KDM6A inducible construct (KDM6A) transfected MIA PaCa-2 cells treated with vehicle control (Ctrl) or Doxycycline (Dox 50 ng/mL) to induce KDM6A expression. GAPDH was used as the reference gene. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001; unpaired t test. chr, chromosome.
KDM6A Deficiency Activates a Noncanonical p38-Dependent Activin A Pathway
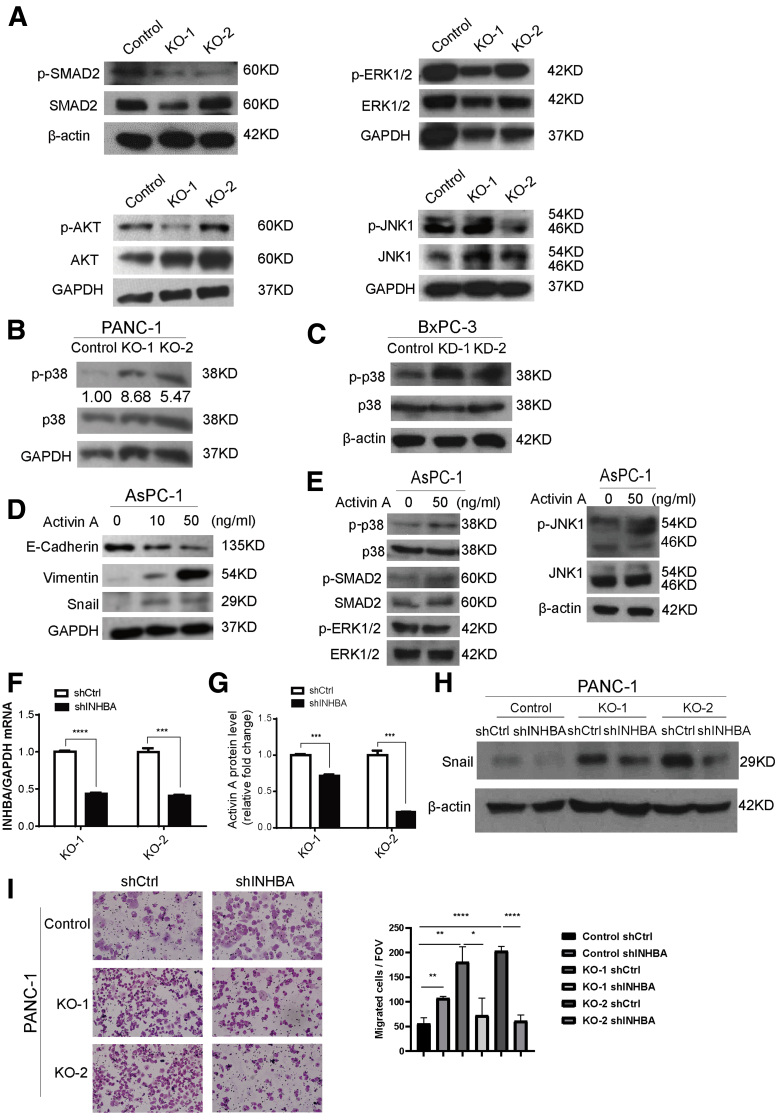
There are 2 activin A signaling cascades: canonical and noncanonical.20 The canonical pathway of activin A signals through Mothers against decapentaplegic homolog (SMAD) 2/3 proteins, similar to the canonical TGF-β pathway. The noncanonical pathway can be mediated by Phosphoinositide 3-kinase (PI3K)/Protein kinase B (AKT), extracellular-signal-regulated kinase (ERK), c-Jun N-terminal kinases (JNK), or p38 mitogen-activated protein kinase (MAPK) signaling. To determine which pathway was activated by KDM6A-induced activin A expression, we surveyed the phosphorylation of the known main regulatory kinases related to the activin A pathway. We found no consistent alterations in p-SMAD2, p-AKT, p-JNK, or p-ERK1/2 levels in KDM6A-KO cells (Figure 7A). However, an increase in p-p38 MAPK was observed in both KDM6A-KO and knockdown cells (Figure 7B and C), supporting the hypothesis that KDM6A signals through a noncanonical p38 MAPK-dependent activin A pathway.
Figure 7.
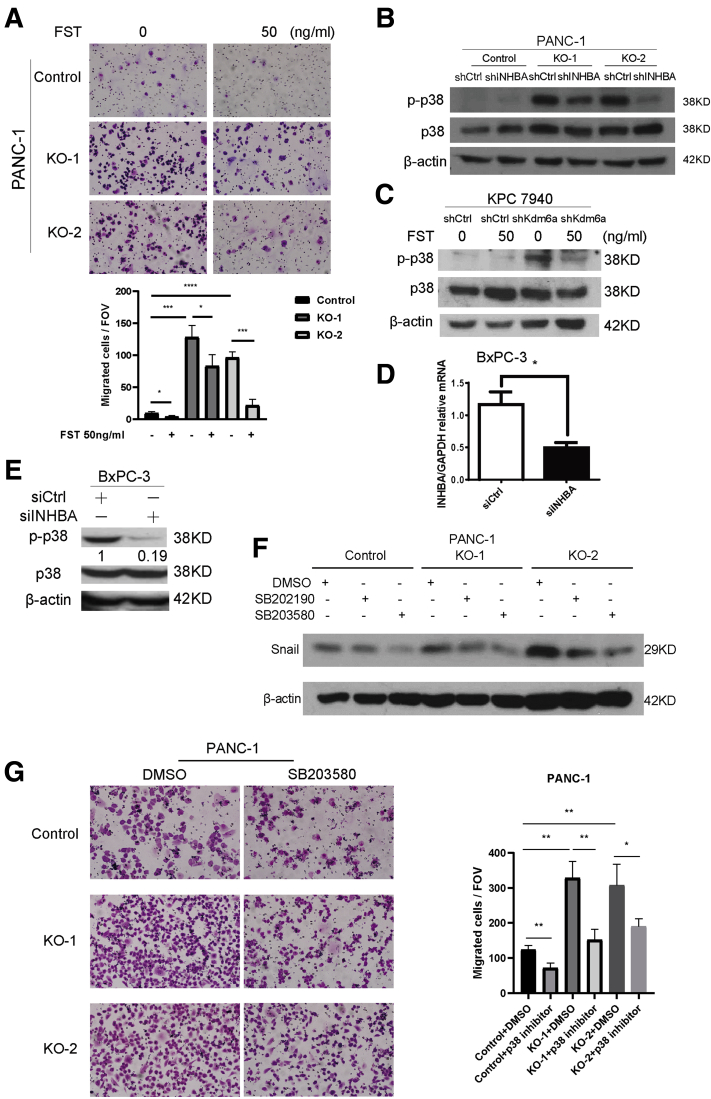
Loss of KDM6A activates a noncanonical p38 MAPK-dependent activin A pathway. (A) Western blot of p-SMAD2, SMAD2, p-ERK1/2, ERK1/2, p-AKT, AKT, p-JNK1, and JNK1 in control and KDM6A-KO PANC-1 cells. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. (B) Western blot of p-p38 and p38 in KDM6A-KO and control PANC-1 cells and (C) KDM6A short hairpin (sh) RNA knockdown or control BxPC-3 cells. GAPDH and β-actin were used as loading controls, respectively. (D) Western blot of E-cadherin, Vimentin, and Snail in AsPC-1 cells treated with vehicle control, or 10 or 50 ng/mL of activin A for 6 hours. GAPDH was used as a loading control. (E) Western blot of p-p38, p38, p-SMAD2, SMAD2, p-ERK1/2, ERK1/2, p-JNK1, and JNK1 in AsPC-1 cells treated with vehicle control or 50 ng/mL of activin A for 6 hours. β-actin was used as a loading control. (F) Real-time reverse-transcription polymerase chain reaction of INHBA in KDM6A-KO PANC-1 cells transfected with control or INHBA shRNA. GAPDH was used as a reference gene. (G) Relative activin A protein levels in condition media from cultured KDM6A-KO PANC-1 cells transfected with either control or INHBA shRNA measured by enzyme-linked immunosorbent assay. (H) Western blot of Snail in the control and 2 KDM6A-KO PANC-1 cell clones transfected with either control or INHBA shRNA. β-actin was used as loading control. (I) Representative images and quantifications of migrated control and KDM6A-KO PANC-1 cells transfected with either control or INHBA shRNA. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001, unpaired t test. Ctrl, control; FOV, field of view; mRNA, messenger RNA.
To determine whether the protumoral and pro-EMT effect of KDM6A loss is mediated by activin A, we first treated PDAC cells with activin A. Indeed, activin A treatment induced a dose-dependent expression reduction of epithelial marker E-cadherin and increased the expression of mesenchymal markers, Vimentin and Snail (Figure 7D), consistent with the EMT process. This transdifferentiation was accompanied by an increase in p-p38 levels, but not in p-SMAD2, p-ERK1/2, or p-JNK levels (Figure 7E), resembling what was observed in KDM6A-deficient cells (Figures 5B and 7A–C). To further confirm that the protumoral function of KDM6A is activin A–dependent, we knocked down INHBA in PDAC cells (Figure 7F and G) and asked whether the transdifferentiation and enhanced cell migration induced by KDM6A loss would be attenuated. Indeed, knockdown of INHBA led to decreased mesenchymal marker Snail expression and reduced cell migration to a level that was comparable with the control cells (Figure 7H and I). In addition, pharmacologic inhibition of activin A using follistatin, a natural activin A antagonist, also significantly attenuated cell migration in both control and KDM6A-knockout PANC-1 cells (Figure 8A). There was also a slight decrease in Snail protein level in the control cells upon INHBA knockdown (Figure 7H). Although it is unclear why INHBA knockdown control cells have increased migration, the overall data support the EMT-promoting function of activin A in both control and KDM6A-KO cells. Mechanistically, activin A knockdown inhibited p38 phosphorylation induced by KDM6A loss (Figure 8B). Similarly, inhibition of activin A by follistatin attenuated p38 phosphorylation induced by KDM6A loss (Figure 8C). In addition, we confirmed this finding in a different PDAC cell line (Figure 8D and E). To assess whether the p38 MAPK pathway is the predominant pathway that mediates KDM6A function in PDAC cells, we used p38 inhibitors, SB202190 and SB203580, to treat control and KDM6A-KO PDAC cells and investigated the EMT process and tumor cell migration. Our data showed that the inhibition of p38 attenuated the EMT process and tumor cell migration in KDM6A-KO PDAC cells to the level of control cells (Figure 8F and G), supporting the p38 MAPK pathway as the dominant pathway that mediates KDM6A–activin signaling in PDAC cells. These results support that activin A–induced p38 pathway activation is most likely the primary signaling that mediates mesenchymal cell identity and cell migration in KDM6A-deficient PDAC cells.
Figure 8.
The activin A–induced p38 pathway is most likely themajorsignalingthatpromotes mesenchymal cell identity and cell migration in KDM6A-deficient PDAC cells. (A) Representative images and quantifications of migrated control and KDM6A-KO PANC-1 cells treated with either vehicle control or 50 ng/mL follistatin (FST). (B) Western blot of p-p38 and p38 in control and KDM6A-KO PANC-1 cells transfected with either control or INHBA short hairpin (sh) RNA. β-actin was used as a loading control. (C) Western blot of p-p38 and p38 in control and KDM6A-knockdown murine KPC 7940 cells treated with either vehicle control or 50 ng/mL FST for 1 hour. β-actin was used as a loading control. (D) Real-time reverse-transcription polymerase chain reaction of INHBA and (E) Western blot of p-p38 and p38 in KDM6A shRNA knockdown BxPC-3 cells transfected with control or INHBA small interfering (si) RNA for 24 hours. Quantification of p-p38 immunoblot band was shown relative to control. (F) Control and KDM6A-KO PANC-1 cells were treated with 10 μmol/L p38 inhibitors, SB202190 or SB203580, or vehicle control dimethyl sulfoxide (DMSO) for 4 days followed by Western blot analysis of Snail. β-actin was used as a loading control. (G) Control and KDM6A-KO PANC-1 cells were treated with 10 μmol/L p38 inhibitor SB203580 or vehicle control DMSO for 4 days followed by Transwell migration assays. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001, unpaired t test. Ctrl, control; FOV, field of view; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Loss of KDM6A Promotes an Aggressive Subtype of PDAC With Enhanced Tumor Growth and Metastasis In Vivo
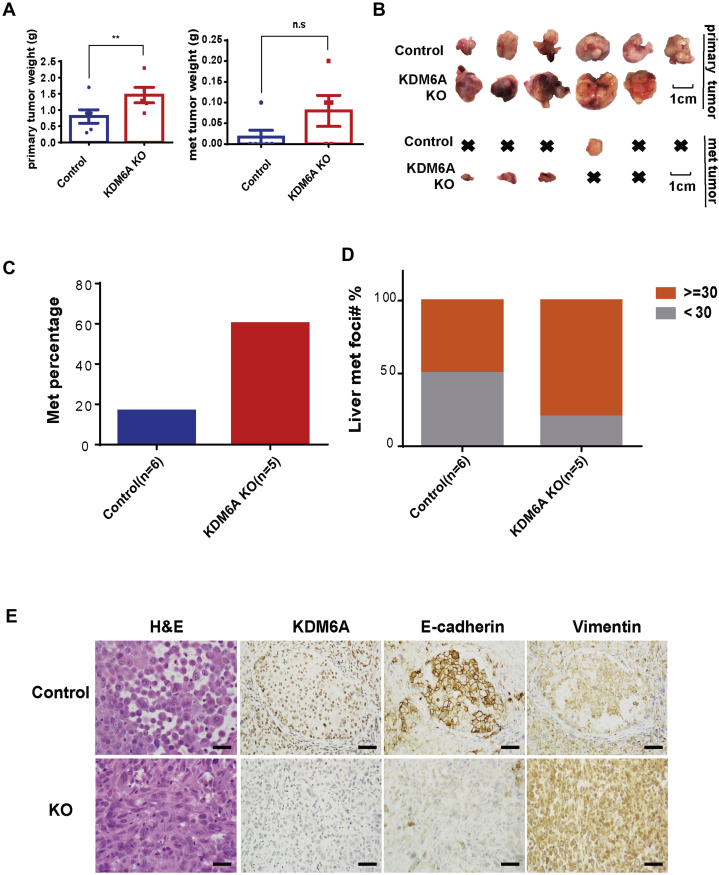
To determine whether the protumoral role of KDM6A loss persists in vivo, we first injected KDM6A-deficient and wild-type human PDAC cells into mouse pancreata. KDM6A-deficient tumors developed into larger primary and metastatic tumors compared with KDM6A wild-type tumors (Figure 9A and B). Furthermore, a higher percentage of animals had muscle metastases and metastatic foci in the liver in the KDM6A-KO group compared with the control group (Figure 9C and D). These findings were consistent with our in vitro observation that loss of KDM6A had a protumor and prometastasis function. To determine whether loss of KDM6A also promoted mesenchymal identity in vivo, H&E and IHC analyses of hallmark epithelial and mesenchymal markers were performed. Confirming our in vitro data, KDM6A-deficient tumors had an undifferentiated sarcomatoid appearance with loss of the epithelial marker E-cadherin and gain of the mesenchymal marker Vimentin (Figure 9E). These data support our hypothesis that loss of KDM6A promotes an aggressive subtype of PDAC with increased tumor growth and metastasis.
Figure 9.
Loss of KDM6A promoted PDAC growth and metastasis in an orthotopic xenograft mouse model. (A) Quantifications of primary and metastatic (met) tumor weight in an orthotopic xenograft mouse model using 106 control or KDM6A-KO PANC-1 cells mixed 1:1 with Matrigel and injected into the pancreatic tail. ∗∗P < .01; unpaired t test. (B) Gross pictures of primary and metastatic tumors from control or KDM6A-KO PANC-1 cells in the orthotopic xenograft mouse model. x, no tumor. Scale bar: 1 cm. (C) Percentage of xenograft mice injected with control or KDM6A-KO PANC-1 cells that showed metastasis. (D) Percentage of xenograft mice injected with control or KDM6A-KO PANC-1 cells with fewer than 30 or 30 or more liver metastatic foci. (E) Representative images of H&E and IHC staining of KDM6A, E-cadherin, and Vimentin in control and KDM6A-KO PANC-1 xenograft tumors. Scale bar: 50 μm.
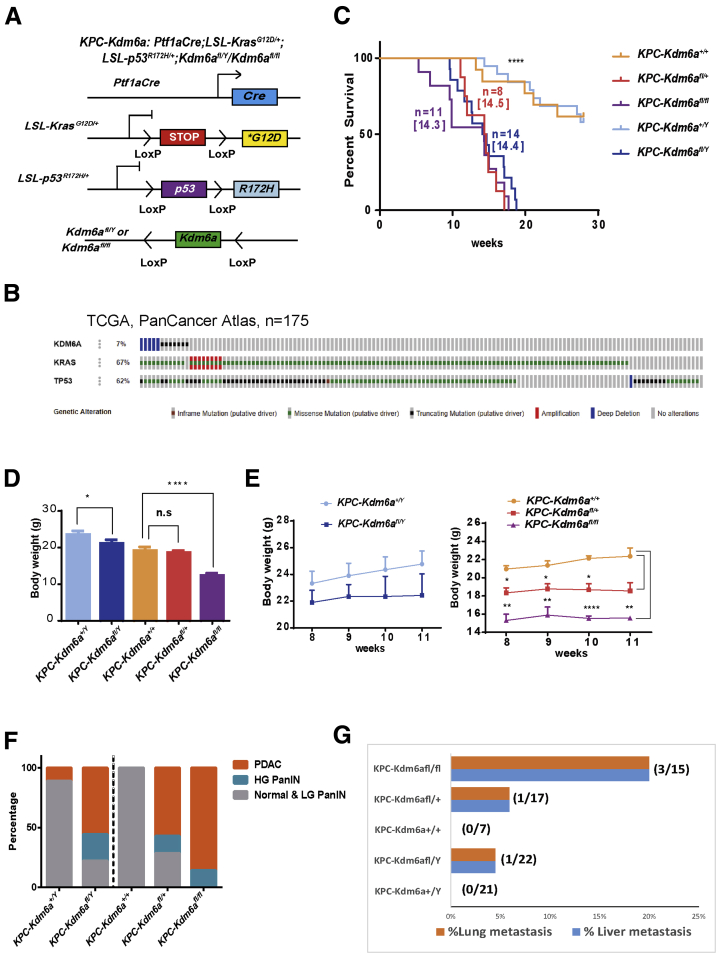
To investigate whether KDM6A is a tumor suppressor in a genetically engineered PDAC mouse model, we crossed Kdm6afl/fl mice with a well-established genetically engineered PDAC mouse model, namely the Ptf1aCre;LSL-KrasG12D/+;LSL-p53R172H/+ (KPC) mice, to generate a pancreas-specific Kdm6a-KO PDAC mouse model (KPC-Kdm6afl/fl or KPC-Kdm6afl/Y) (Figure 10A).21,22 We decided to use the KPC mouse model because the majority of human PDACs carrying KDM6A mutations coincided with both KRAS and TP53 mutations according to TCGA database (Figure 10B). Ptf1aCre, Kdm6afl/fl mice did not have any abnormalities in pancreas weight, pancreas histology, or body weight (data not shown), as previously described.15 However, KPC-Kdm6afl/fl, KPC-Kdm6afl/+, and KPC-Kdm6afl/Y mice had significantly shorter lifespans compared with KPC mice (Figure 10C). KPC-Kdm6afl/fl and KPC-Kdm6afl/Y mice also had a lower body weight, especially female KPC-Kdm6afl/fl mice, compared with KPC mice (Figure 10D and E). KPC mice were known to develop PDAC at approximately 16–20 weeks of life.23,24 In comparison, in mice younger than age 12 weeks, more KPC-Kdm6afl/fl, KPC-Kdm6afl/+, and KPC-Kdm6afl/Y mice developed PDAC than KPC mice (Figure 10F). KPC-Kdm6afl/fl mice developed PDAC at week 12.3 ± 1.3 (n = 11) and KPC-Kdm6afl/Y mice at week 14.3 ± 0.87 (n = 14) compared with week 18.1 ± 1.8 (n = 6) in KPC mice. All KPC-Kdm6afl/fl mice developed high-grade PanIN and PDAC by the age of 12 weeks, while almost all KPC mice had low-grade PanIN (PanIN1–2) at the same age (Figure 10F). Moreover, we observed the most lung and liver metastases in female KPC-Kdm6afl/fl mice (20%; 3 of 15), less in male KPC-Kdm6afl/Y mice (4.5%; 1 of 22) and female KPC-Kdm6afl/+ mice (5.9%; 1 of 17) compared with the same age KPC mice, which had no lung or liver metastases (Figure 10G).
Figure 10.
Loss of KDM6A promotes an aggressive subtype of PDAC with accelerated tumor growth and metastasis in mice. (A) Schematic diagram of Ptf1aCre;LSL-KrasG12D/+;LSL-p53R172H/+;Kdm6afl/Y/ Kdm6afl/fl (KPC-Kdm6a) mouse model. (B) Sequencing data for human PDACs (N = 175) were queried from TCGA PanCancer Atlas database and PDACs with KDM6A, KRAS, and TP53 mutations were highlighted by various colored bars. (C) Kaplan–Meier plot showing the survival of male and female KPC-Kdm6a mice compared with KPC mice. Median survival is shown in brackets. n, number of mice. Log-rank (Mantel–Cox) test. (D) Body weight of male and female KPC-Kdm6a mice. (E) Body weight of male (left) and female (right) KPC-Kdm6a mice at 8, 9, 10, and 11 weeks. (F) Percentage of mouse pancreata with normal or low-grade (LG) PanIN (PanIN1 and 2), high-grade (HG) PanIN (PanIN3), and PDAC lesions in male and female KPC-Kdm6a mouse models. (G) Percentage of mice with lung and liver metastases in male and female KPC-Kdm6a mouse models younger than 23 weeks old. Fisher exact test. ∗P < .05, ∗∗P < .01, and ∗∗∗∗P < .0001.
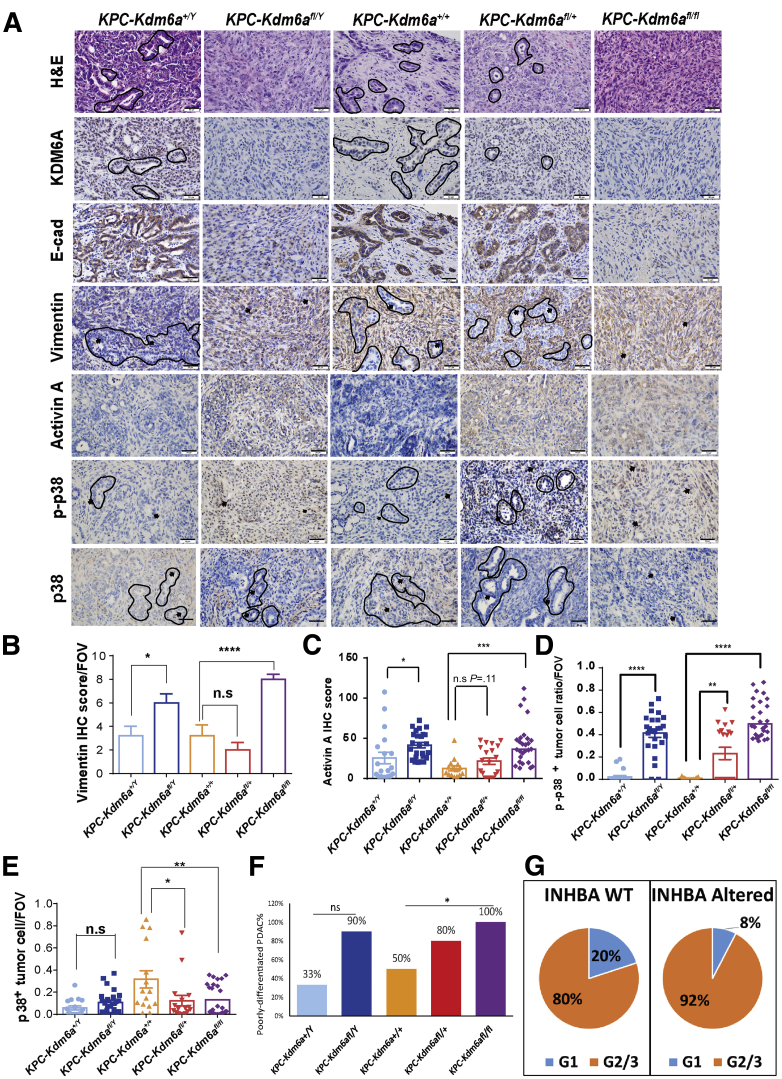
We then analyzed the histology and immunohistochemical markers in the pancreatic tumors from these mice. Loss of KDM6A (Figure 11A, second row) led to the development of an aggressive undifferentiated sarcomatoid carcinoma (Figure 11A, top row). In addition, consistent with our in vitro and xenograft models, KDM6A-deficient tumors lost the epithelial marker, E-cadherin (Figure 11A, third row), and gained the mesenchymal marker, Vimentin (Figure 11A, fourth row, and B), indicating the EMT process. In contrast, pancreatic tumors from KPC or KPC-Kdm6afl/+ mice were positive for E-cadherin and negative for Vimentin (Figure 11A and B). Activin A and p-p38 levels also were increased in KDM6A-deficient tumors (Figure 11A, bottom 3 rows, and C–E). Histologically, 100% of KPC-Kdm6afl/fl mice and 90% of KPC-Kdm6afl/Y mice developed poorly differentiated or undifferentiated PDAC, compared with 33% and 50% of KPC male and female mice, respectively (Figure 11F). To investigate if INHBA alteration also was associated with less-differentiated PDAC in human beings, we surveyed the TCGA database. Consistent with our hypothesis, INHBA-altered PDACs are composed of a higher percentage of G2/G3 (moderately or poorly differentiated) tumors and less G1 (well-differentiated) tumors compared with INHBA wild-type PDACs (92% G2/3 in INHBA-altered PDACs vs 80% G2/3 in INHBA-wild type PDACs) (Figure 11G). These results further support our hypothesis that loss of KDM6A promotes an aggressive subtype of PDAC and shortens survival by modulating cancer cell identity via the noncanonical p38 MAPK-dependent activin A pathway.
Figure 11.
Loss of KDM6A induces poorly or undifferentiated type PDACs with increased activin A expression and p38 phosphorylation in mice pancreata. (A) Representative images of H&E and IHC staining of KDM6A, E-cadherin, Vimentin, activin A, p-p38, and p38 in PDAC samples from male and female KPC-Kdm6a mouse models. A few differentiated PDAC tubules are outlined by black lines. Arrows point to representative PDAC cell nuclei. Scale bar: 50 μm. (B–E) Quantifications of Vimentin, activin A, p-p38, and p38 IHC staining in PDAC samples of male and female KPC-Kdm6a mouse models. Unpaired t test. Number of mice in each group: KPC-Kdm6a+/Y, 4; KPC-Kdm6afl/Y, 5; KPC-Kdm6a+/+, 3; KPC-Kdm6afl/+, 4; and KPC-Kdm6afl/fl, 7. (F) Mice PDACs were classified as either poorly differentiated or well/moderately differentiated according to histology. Percentages of poorly differentiated PDAC in each group are shown. (G) Histology grade data of human PDACs (N = 175) were queried from TCGA PanCancer Atlas database. Percentages of G1 and G2/3 PDACs in INHBA wild-type (WT) or altered tumors are shown. Fisher exact test and unpaired t test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. FOV, field of view.
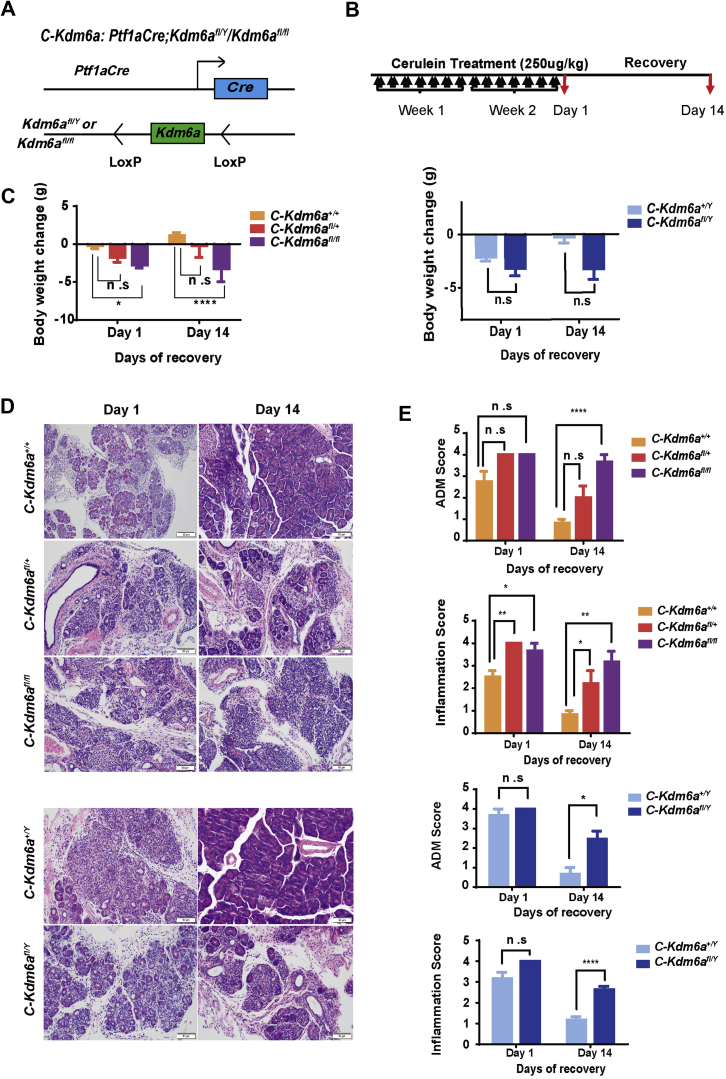
KDM6A Deficiency Alone Delays Recovery From Cerulein-Induced Chronic Pancreatitis
Chronic pancreatitis is a known susceptibility factor for PDAC and, in the background of oncogenic KRAS, can accelerate the progression to PanIN and PDAC.25,26 Although KDM6A deficiency alone did not lead to any abnormal phenotype in the pancreata of our Kdm6a-KO mice, we speculate that it may contribute to an attenuated recovery from injuries such as pancreatitis, and thus increased susceptibility to PDAC. We tested this hypothesis using our Kras wild-type, pancreas-specific Kdm6a-KO mice, namely Ptf1aCre;Kdm6afl/fl, or Ptf1aCre;Kdm6afl/Y (C-Kdm6afl/fl or C-Kdm6afl/Y) (Figure 12A), in a well-established, cerulein-induced, chronic pancreatitis model.27 Mice were treated with cerulein for 2 weeks and then allowed to recover for 1 and 14 days (Figure 12B). There was a significant and persistent weight loss in female C-Kdm6afl/fl mice at both day 1 and day 14 of recovery compared with the wild-type Ptf1aCre;Kdm6a+/+ (C) mice (Figure 12C). Heterozygous female C-Kdm6afl/+ mice and male C-Kdm6afl/Y mice also lost more weight than C mice, but the difference was not statistically significant. Histologically, all pancreata showed marked inflammation and extensive acinar-to-ductal metaplasia (ADM), a precursor for PDAC, on day 1 of recovery (Figure 12D). However, pancreata from female C-Kdm6afl/+ and C-Kdm6afl/fl mice contained even more inflammation than C mice on day 1 (Figure 12E). No significant difference in ADM was observed among groups on day 1. After recovering for 14 days, the majority of the pancreata from C animals completely recovered with regeneration of acini and minimal residual inflammation and ADM (Figure 12D and E). However, both male and female Kdm6a-deficient mice, including female heterozygous Kdm6a+/fl mice, failed to recover with persistent inflammation and ADM on day 14 (Figure 12D and E). These findings support our hypothesis that KDM6A deficiency delays recovery from chronic pancreatitis and promotes persistent ADM and inflammation, which may accelerate KRAS-induced pancreatic tumorigenesis.
Figure 12.
KDM6A deficiency delays recovery from cerulein-induced chronic pancreatitis. (A) Schematic diagram of Ptf1aCre;Kdm6afl/Y/Kdm6afl/fl (C-Kdm6a) mice. (B) Experimental design for chronic pancreatitis induction in C-Kdm6a mice. Mice were injected intraperitoneally with 250 μg/kg cerulein twice a day for 2 consecutive weeks followed by recovery for 1 and 14 days. (C) Body weight changes in male (right) and female (left) C-Kdm6a mice on day 1 and day 14 of recovery. (D) Representative images of H&E staining. Scale bar: 50 μm. (E) Quantification of ADM and inflammation in male and female C-Kdm6a mice pancreata on day 1 and day 14. At least 6 mice were analyzed in each group. ∗P < .05, ∗∗P < .01, and ∗∗∗∗P < .0001; unpaired t test.
Discussion
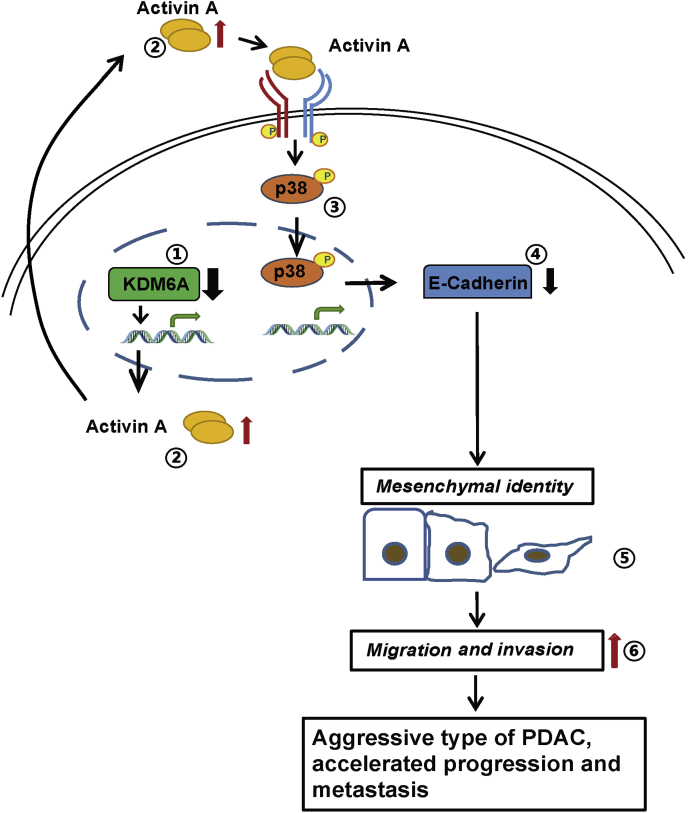
KDM6A is one of the most frequently mutated epigenetic genes in PDAC.7,8 However, its role in PDAC development is not fully understood. Earlier studies by Andricovich et al15 showed that loss of KDM6A induced a gender-specific, squamous-like pancreatic cancer through activation of superenhancers regulating ΔNp63, MYC, and RUNX3 oncogenes using the Ptf1aCre;LSL-KrasG12D/+ (KC) mouse model. However, which signaling pathway mediates KDM6A function in PDAC development remains elusive. Furthermore, the majority of KDM6A-mutated human PDACs also carry TP53 mutations in addition to KRAS mutations (TCGA database) (Figure 10B). Whether KDM6A loss accelerates PDAC progression in the presence of both KRAS and TP53 mutations is not known. In this study, we generated pancreas-specific Kdm6a-KO mice in the background of the well-accepted KPC PDAC mouse model carrying both KRAS and TP53 mutations. In addition, we discovered that KDM6A loss alone attenuated acinar regeneration and recovery from chronic pancreatitis–induced ADM. Our findings strongly support the tumor-suppressive role of KDM6A in pancreas tumorigenesis and progression. In addition, we have unveiled the mechanism by which loss of KDM6A promoted the development of an aggressive, undifferentiated subtype of PDAC, primarily mediated by a noncanonical p38 MAPK-dependent activin A pathway (Figure 13). However, we cannot entirely exclude the role of the canonical activin A pathway in KDM6A signaling without further investigation. Similar to the previous study,15 our results also suggest that the tumor-suppressive role of KDM6A in PDAC is likely enzyme-independent because there were no significant changes in the global H3K27me3 level. Because KDM6A is a component of the COMPASS (Complex Proteins Associated with Set1) -like complex, which plays an essential role in regulating promoters and enhancers, we speculate that KDM6A may cooperate with the other components of the COMPASS-like complex to regulate INHBA transcription. Supportive evidence of this hypothesis came from our unpublished data on another component of the complex, KMT2D, which can regulate INHBA expression as well.
Figure 13.
Schematic model showing the tumor-suppressive role of KDM6A in PDAC progression. KDM6A deficiency leads to the increased expression of activin A. Activin A activates a noncanonical p38 MAPK pathway, which induces mesenchymal identity, tumor sphere formation, cell migration, and invasion in PDAC cells. This signaling promotes the formation of an aggressive type of PDAC with accelerated progression and metastasis.
Our findings are different from the observations made in the previous KC mouse model. There was no significant dosage effect of Kdm6a allele deletion on survival in our KPC mouse model. The female heterozygous Kdm6a-KO KC mice lived 2–3 times longer than the homozygous Kdm6a-KO KC mice.15 In contrast, the median survival rate of our female homozygous Kdm6a-KO KPC mice was similar to the heterozygous Kdm6a-KO mice, suggesting that loss of even 1 copy of Kdm6a in KPC mice is sufficient to accelerate PDAC progression. We postulate the following possible reasons for these differences. First, the p53 mutation may contribute to the diminished difference between homozygous and heterozygous mice. We introduced a p53 mutation into the KC mouse model because of its closer resemblance to the mutation pattern observed in human PDAC samples. Second, our inclusion criteria for the survival curve may be different from the Andricovich et al study.15 We included mice that were either dead or had disabilities and more than 15% weight loss within 1 week into the survival curve analysis. However, we did observe a Kdm6a dose-dependent difference in ADM and inflammation levels in our chronic pancreatitis model without p53 and Kras influence.
Similar to the KC model,15 we also observed gender differences in metastasis and recovery from chronic pancreatitis. More female Kdm6a-KO KPC mice developed lung and liver metastasis compared with male Kdm6a-KO KPC mice (20% in female vs 4.5% in male). Female Kdm6a-KO mice also recovered more slowly than male Kdm6a-KO mice from chronic pancreatitis. However, we did not observe gender differences in PDAC progression or survival as described in the KC mouse model. Again, these differences could be explained by the addition of the p53 mutation in our model.
The reversible EMT process is one of the most important regulators of cell plasticity and metastasis.28, 29 We found that the EMT pathway is one of the top pathways that was up-regulated in KDM6A-deficient cells. This result is consistent with the RNA sequencing data of cell lines established from Kdm6a-KO KC mice pancreata.15 The TGF-β pathway is a well-known pathway that regulates EMT.28 Activin A is a member of the TGF-β superfamily, which plays a critical role in embryonic pancreatic development.30 Activin A also participates in a variety of biological processes, from cell differentiation to cell migration and wound healing.20, 29 Similar to TGF-β, activin A was shown to have both tumor-suppressive and oncogenic roles in cancer depending on the specific cell type and context.18 In certain tumor types, including lung and head and neck cancers, activin A expression is associated with proliferation, invasion, and poor prognosis.18 In contrast, in breast and prostate cancers, activin A plays a tumor-suppressive role.18 The exact role of activin A in PDAC remains elusive. High plasma activin A levels are associated with shorter survival periods in PDAC patients.19 In a xenograft mouse model, overexpression of activin A promotes PDAC growth and cachexia,31 supporting its critical role in PDAC progression. However, blocking the activin pathway by a soluble form of activin-receptor IIB and Anaplastic lymphoma kinase 4 (ALK4) KO in the very early stage of PDAC development in KC mice appeared to accelerate the formation of ADM and PanIN lesions. In another study by Qiu et al, ALK4 knockout in KC mice accelerated intraductal papillary mucinous neoplasm development, but not PanIN lesions.32 We have found activin A to be one of the top up-regulated genes in the EMT pathway upon KDM6A loss. We then discovered that KDM6A loss induced a noncanonical p38 MAPK-mediated activin A pathway in PDAC, which promoted mesenchymal identity, tumor sphere formation, cell migration and invasion in vitro, and induced the formation of an aggressive undifferentiated subtype of PDAC in KPC mice accompanied by increased metastasis and shorter survival. These findings strongly support that the KDM6A–activin axis plays a tumor-promoting role in PDAC. In addition, we postulate that activin A may contribute to significant weight loss in Kdm6a-KO KPC mice, possibly related to cachexia as previously described.19, 33, 34
In conclusion, we showed that loss of KDM6A in the pancreas promoted an aggressive undifferentiated subtype of PDAC with mesenchymal differentiation primarily by activating a noncanonical p38 MAPK-mediated activin A pathway. Furthermore, KDM6A deficiency alone delayed recovery from chronic pancreatitis with persistent ADM and inflammation. Blocking activin A attenuated the protumoral function of KDM6A deficiency. These results not only showed a novel signaling pathway of KDM6A in PDAC development, but also identified activin A as a potential target for the treatment of PDACs with KDM6A mutations.
Materials and Methods
Cell Lines
Human PDAC cell lines PANC-1, MIA PaCa-2, and UM28 (gifts from Mats Ljungman, University of Michigan) were cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum. BxPC-3 and AsPC-1 PDAC cells were cultured in RPMI supplemented with 10% fetal bovine serum. All cells were cultured at 37°C with 5% CO2. The KDM6A-KO PANC-1 cell line was generated using Edit-R Lentiviral Cas9 nuclease vectors with synthetic CRISPR RNAs (Dharmacon, Lafayette, CO), followed by single clone isolation and expansion. The KDM6A-overexpressed MIA PaCa-2 cell line was generated using the Tet-3G inducible expression system (Takara Bio, San Jose, CA) according to the manufacturer’s instructions. Briefly, KDM6A gene was cloned from pCS2 vector (kindly shared by Dr Kai Ge) into pLVX-TRE3G vector (Takara Bio, San Jose, CA). MIA PaCa-2 cells then were co-transduced with Lentiviral pLVX-TRE3G-KDM6A and pLVX-Tet3G particles and cultured in selection medium to select stable colonies. MIA PaCa-2 cells co-transduced with empty lentiviral pLVX-TRE3G and pLVX-Tet3G particles were used as a negative control. INHBA RNA interference (RNAi) iLenti construct (Applied Biological Materials Inc., Richmond, BC, Canada) and Lipofectamine 2000 (ThermoFisher, Waltham, MA) were used to knockdown INHBA according to the manufacturer’s instructions. Stable knockdown of KDM6A was established by transducing GipZ Lentivirus Human KDM6A short hairpin RNA into PDAC cells followed by sorting GFP (green fluorescent protein)-positive cells and expanding single-cell clones. Knockdown of KDM6A was achieved by direct transfection of KDM6A small interfering RNAs into cells by the Lipofectamine RNAiMAX reagent (ThermoFisher, Waltham, MA) according to the manufacturer’s instructions.
Clinical Samples
The study was approved by the Institutional Review Board at the University of Michigan (number: HUM00098128). Patients with pancreas resections for pancreatitis, cystic neoplasms, or PDAC from 2002 to 2015 at the University of Michigan Health System were included in the study.
TMAs
All H&E-stained slides were reviewed, and diagnoses were confirmed and corresponding areas were carefully selected and marked. Duplicated 1-mm diameter tissue cores from a total of 213 patient tissue samples were selectively punched and transferred to recipient tissue array blocks. Five TMAs were set up according to a standard protocol, as previously described.35 H&E and IHC staining was performed on each TMA block using standard protocols.
Bru-seq Analysis
Bru-seq was performed as previously described.17 Briefly, PANC-1 cells were incubated in media containing bromouridine at a final concentration of 2 mmol/L for 30 minutes at 37°C to label nascent RNA. Cells then were lysed directly in TRIzol (ThermoFisher, Waltham, MA), and total RNA was isolated. Bru-labeled RNA was immunoprecipitated using antibromodeoxyuridine antibody, followed by the preparation of strand-specific complementary DNA libraries with the TruSeq kit (Illumina, San Diego, CA) and deep sequencing using the Illumina sequencing platform as previously described.17,36,37 Gene Set Enrichment Analysis was performed as previously described.38
RNA Isolation and Quantitative Real-Time Reverse-Transcription Polymerase Chain Reaction
Total RNAs were extracted using the RNeasy Mini Kit (QIAGEN, Germantown, MD). Reverse transcription of RNA was performed using the SuperScript III First-Strand Synthesis kit (ThermoFisher, Waltham, MA). Quantitative real-time polymerase chain reaction was performed with SYBR Green (ThermoFisher, Waltham, MA) reagents in MicroAmp Optical 96-well reaction plates (ThermoFisher, Waltham, MA). Primers were designed by Primer-BLAST using the National Center for Biotechnology Information (NCBI) primer designing tool and synthesized by Integrated DNA Technologies (Coralville, IA).
Western Blot Analysis
Cells were harvested with NP-40–based whole-cell lysis buffer (50 mmol/L Tris, pH 8.0, 150 mmol/L NaCl, 2 mmol/L EDTA, 1 mmol/L phenylmethylsulfonyl fluoride, 1× proteinase inhibitor, and 1.5% NP-40). Protein concentrations were measured using the Bradford assay. Protein samples were heated and separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis or Tris–acetate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. After blocking in phosphate-buffered saline/Tween-20 containing 5% nonfat milk, the membranes were incubated with primary antibodies (Table 2) overnight at 4°C, followed by incubation with peroxidase-conjugated secondary antibodies for 1 hour. The proteins were visualized using an ECL (Enhanced chemiluminescence) detection kit.
Table 2.
Information of Reagents and Antibodies
| Reagents or antibodies | Company (catalog number, concentration) |
|---|---|
| Activin A | PEPROTECH (Rocky Hill, NJ, #120-14P, 10 ng/mL) |
| Follistatin | PEPROTECH (Rocky Hill, NJ, #120-13) |
| Cerulein | Sigma (St. Louis, MO, #C9026) |
| Lipofectamine 2000 | ThermoFisher (Waltham, MA, #11668030) |
| Puromycin | Sigma (St. Louis, MO, #P8833) |
| Human/Mouse/Rat Activin A Quantikine ELISA Kit | R&D Systems (Minneapolis, MN, #DAC00B) |
| E-cadherin | Cell Signaling (Danvers, MA, #3195, 1/1000) |
| MMP2 | Cell Signaling (Danvers, MA, #40994, 1/1000) |
| SNAIL | Cell Signaling (Danvers, MA, #3879, 1/1000) |
| GAPDH | Cell Signaling (Danvers, MA, #5174, 1/1000) |
| β-actin | Sigma-Aldrich (St. Louis, MO, #A5441,1/2500) |
| Vimentin | Santa Cruz (Dallas, TX, #SC66002, 1/500) |
| H3K4me1 | Cell Signaling (Danvers, MA, #5326s, 1/1000) |
| H3K4me3 | Cell Signaling (Danvers, MA, #9751s, 1/1000) |
| H3K27me3 | Cell Signaling (Danvers, MA, #9733, 1/1000) |
| H3K27ac | Abcam (Cambridge, United Kingdom, #ab4729) |
| H3 | Cell Signaling (Danvers, MA, #4499, 1/1000) |
| KDM6A | Cell Signaling (Danvers, MA, #33510s, 1/1000) |
| p38 | Cell Signaling (Danvers, MA, #8690, 1/1000) |
| p-p38 | Cell Signaling (Danvers, MA, #4511, 1/1000) |
| ERK | Cell Signaling (Danvers, MA, #4691, 1/1000) |
| p-ERK | Cell Signaling (Danvers, MA, #2965, 1/1000) |
| Akt | Cell Signaling (Danvers, MA, #4691, 1/1000) |
| p-Akt | Cell Signaling (Danvers, MA, #2965, 1/1000) |
| JNK1 | Cell Signaling (Danvers, MA, #3708, 1/1000) |
| p-SAPK/JNK | Cell Signaling (Danvers, MA, #4668, 1/1000) |
| SMAD2 | Cell Signaling (Danvers, MA, #3103, 1/1000) |
| p-SMAD2 | Cell Signaling (Danvers, MA, #3108s, 1/500) |
| SMAD3 | Cell Signaling (Danvers, MA, #9523s, 1/1000) |
| p-SMAD3 | Cell Signaling (Danvers, MA, #9520s, 1/500) |
GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Transwell Migration and Invasion Assay
In vitro Transwell migration and invasion assays were performed using a 24-well plate with 8-μm pore size chamber inserts. PANC-1 cells (3 × 104) or BxPC-3 cells (8 × 103) in serum-free media were seeded onto a noncoated membrane for migration assays or a Matrigel (ThermoFisher, Waltham, MA)-coated membrane for invasion assay in the upper chamber. Complete growth medium (500 μL) was added to the lower chamber. After incubation for 24–48 hours at 37°C, the cells that invaded or migrated through the membrane were fixed and stained with the DIFF staining kit (IMEB Inc., San Marcos, CA) and counted in more than 5 random fields.
Three-Dimensional Cell Culture and Invasion Assay
Cells were trypsinized and cultured in a 6-well Ultra-Low Attachment Plate (ThermoFisher, Waltham, MA) overnight to form spheroids. Growth media containing 2 mg/mL type I collagen was added into 8-well chamber slides and left to solidify at room temperature for 30 minutes. The spheroids then were mixed with type I collagen and added to the chamber and incubated at room temperature for 30 minutes before adding culture media into the chambers. The chamber slide was incubated at 37°C for 24 and 48 hours. Images were captured by an inverted microscope.
Tumor Sphere Formation Assay
Single cells (1 × 104) were suspended in tumor sphere culture media X-VIVO 10 Serum-free Hematopoietic Cell Medium (Lonza, Bend, OR) in 6-well Ultra-Low Attachment Plates for 5–7 days. Pictures of the tumor sphere were taken using an inverted microscope.
IHC and H&E Analysis and Scoring
IHC was performed as previously described.39,40 Briefly, formalin-fixed and paraffin-embedded tissues were cut into 5-μm sections and mounted on charged slides. After deparaffinization, antigen retrieval, and blocking, slides were incubated with their respective primary antibodies overnight at 4°C. Slides then were incubated with a peroxidase-labeled secondary antibody, developed with the 3,3′-diaminobenzidine tetra hydrochloride substrate kit, and counterstained with hematoxylin. For E-cadherin and Vimentin IHC score, the intensity of the staining was recorded as follows: 0, negative; 1, weak; 2, moderate; and 3, strong, and the percentage of positive cells was assessed. The final score was calculated as staining intensity × positive cells (%). For activin A IHC, the score was calculated as the reference suggested.41 For p-p38 and p38 IHC, the percentage of positive tumor cells/total tumor cells per field was calculated. ADM and inflammation in pancreatic tissues were scored using a grading scale described previously.42
ELISA
Secreted activin A protein in the culture media after 24 hours of culture was measured with ELISA using the Human/Mouse/Rat Activin A Quantikine ELISA Kit (R&D Systems, Inc., Minneapolis, MN) according to the manufacturer’s instructions.
Mice
All animal studies were approved by the Institutional Animal Care and Use Committee at the University of Michigan. The experimental mice were generated by crossing Kdm6afl/fl mice (stock no. 024177; Jackson Laboratory, Bar Harbor, ME)43 with Ptf1aCre (gift from C. Wright, Vanderbilt University, Nashville, TN),21 and/or LSL-KrasG12D/+ (stock no. 008179; Jackson Laboratory)22,44 and LSL-p53R172H/+ (stock no. 008652; Jackson Laboratory)45 mice. For orthotopic xenograft mouse models, 6- to 8-week-old NOD SCID-Il2rg-/- immunodeficient mice were used (stock no. 005557; Jackson Laboratory).
Statistical Analyses
A 2-tailed unpaired Student t test, an unpaired t test with Welch’s correction, Mann–Whitney, and 1-way analysis of variance with the Turkey multiple comparisons test were used (GraphPad Prism 7.0, San Diego, CA). Statistical significance was defined as P < .05. The results are shown as means ± SEM. The number of independent experiments (N) is indicated in the figure legends.
All reagents and antibodies used are listed in Table 2. RNAi and vector information is listed in Table 3. Primer sequences are provided in Table 4.
Table 3.
RNAi and Vector Construct Information
| Name | Sequence | Company (catalog #) |
|---|---|---|
| tracrRNA | N/A | Dharmacon (Lafayette, CO, #U-002005-05) |
| KDM6A crRNA | TACTGAATTGCTAGGCAGGG | Dharmacon (Lafayette, CO, #CM-014140-04) |
| scramble siRNA | UGGUUUACAUGUCGACUAA | Dharmacon (Lafayette, CO, #D-001210-05) |
| KDM6A siRNA1 | GAGAGUAAUUCACGAAAGA | Dharmacon (Lafayette, CO, #D-014140-02) |
| KDM6A siRNA2 | CAGCACGAAUUAAGUAUUU | Dharmacon (Lafayette, CO, #D-014140-04) |
| INHBA siRNA | GAACGGGUAUGUGGAGAUA | Dharmacon (Lafayette, CO, #D-011701-02) |
| piLenti-siRNA- scramble-GFP | GGGTGAACTCACGTCAGAA | abm (Richmond, BC, Canada, #LV015-G) |
| piLenti-siINHBA-GFP | ACCTCGGAGATCATCACGTTTGCCGAGTC | abm (Richmond, BC, Canada, #i010997) |
| pGipZ-scramble-GFP | N/A | Open Biosystems (Huntsville, AL, #RHS4346) |
| pGipZ-shKDM6A-GFP | N/A | Open Biosystems (Huntsville, AL, #V2LHS_66490) |
GFP, green fluorescent protein; N/A, not applicable; siRNA, small interfering RNA.
Table 4.
Primer Sequences
| Name | Sequence |
|---|---|
| h-CDH1 | Forward: 5’-GGCCTGAAGTGACTCGTAACG-3’ Reverse: 5’-CAGTATCAGCCGCTTTCAGATTT-3’ |
| h-MMP2 | Forward: 5’-TTGATGGCATCGCTCAGATC-3’ Reverse: 5’-TTGTCACGTGGCGTCACAGT-3’ |
| h-SNAIL | Forward: 5’-CAGACCCACTCAGATGTCAA-3' Reverse: 5’-CATAGTTAGTCACACCTCGT-3’ |
| h-VIM | Forward: 5’-CCTTGAACGCAAAGTGGAATC-3’ Reverse: 5’-GACATGCTGTTCCTGAATCTGAG-3’ |
| h-KDM6A | Forward: 5’-CAAGGCTGTTCGCTGCTATG-3’ Reverse: 5’-AGGCAGCATTCTTCCAGTAGTC-3’ |
| h-GAPDH | Forward: 5’-ACCCAGAAGACTGTGGAT-3’ Reverse: 5’-GAGGCAGGGATGATGTTC-3’ |
| h-INHBA | Forward: 5’-GTGAGTGCCCGAGCCATATAG-3’ Reverse: 5’-CATGCGGTAGTGGTTGATGACT-3’ |
| m-INHBA | Forward: 5’-GTGGTGCCAGTCTAGTGCTT-3’ Reverse: 5’-GCAAATGTTGACCTTGCCGT-3’ |
| m-LSL-KrasG12D | Universal: 5’-CGCAGACTGTAGAGCAGCG-3’ Mutant: 5’-CCATGGCTTGAGTAAGTATGC-3’ |
| m-LSL-Trp53 | Forward 1: 5’-TTACACATCCAGCCTCTGTGG-3’ Forward 2: 5’-AGCTAGCCATGGCTTGAGTAAGTCT-3’ Reverse: 5’-CTTGGAGACATAGCCACAGTG-3’ |
| m-KDM6A | Forward: 5’-GCTACTGGGGTGTTTTGAATG-3’ Reverse: 5’-TTTCATAGAACAGTTTCAGGATACC-3’ |
| m-Ptf1aCre | Forward: 5’-CATGCTTCATCGTCGGTCC-3’ Reverse: 5’-GATCATCAGCTACACCAGAG-3’ |
h, human cells or tissues; m, mouse cells or tissues.
Acknowledgments
The authors thank Daniel Long for technical support in helping with mouse tissue embedding and unstained slide preparation, Christopher Wright for sharing Ptf1aCre mice, Kai Ge for sharing KDM6A constructs and antibody, Susanna Maisto for proofreading the manuscript, and all members of the Pancreatic Cancer Research group at the University of Michigan for technical assistance and helpful discussions.
CRediT Authorship Contributions
Zhujun Yi, MD (Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Validation: Lead; Visualization: Lead; Writing – original draft: Lead)
Shanqiao Wei, PhD (Data curation: Equal; Formal analysis: Equal; Investigation: Equal; Methodology: Equal; Writing – review & editing: Equal)
Lin Jin, MD (Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting; Visualization: Supporting; Writing – review & editing: Supporting)
Sivakumar Jeyarajan, PhD (Formal analysis: Equal; Investigation: Equal; Methodology: Equal)
Jing Yang, MD (Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting)
Yumei Gu, MD (Data curation: Supporting; Formal analysis: Supporting; Writing – review & editing: Supporting)
Hong Sun Kim, PhD (Data curation: Supporting; Formal analysis: Supporting; Project administration: Supporting; Writing – review & editing: Supporting)
Shula Schechter, MD (Data curation: Supporting; Formal analysis: Supporting)
Shuang Lu, MD (Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting)
Michelle T Paulsen, BS (Project administration: Supporting; Resources: Supporting)
Karan Bedi, BS (Formal analysis: Supporting)
Ishwarya Venkata Narayanan, BS (Formal analysis: Supporting)
Mats Ljungman, PhD (Conceptualization: Supporting; Resources: Supporting; Supervision: Supporting)
Howard C Crawford, PhD (Resources: Supporting)
Marina Pasca di Magliano, PhD (Resources: Supporting)
Kai Ge, PhD (Resources: Supporting; Writing – review & editing: Supporting)
Yali Dou, PhD (Conceptualization: Supporting; Funding acquisition: Supporting; Resources: Supporting; Supervision: Supporting)
Jiaqi Shi, MD, PhD (Conceptualization: Lead; Data curation: Lead; Formal analysis: Supporting; Funding acquisition: Lead; Investigation: Lead; Methodology: Lead; Project administration: Lead; Resources: Lead; Supervision: Lead; Visualization: Supporting; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported in part by National Institutes of Health grants K08CA234222 (J.S.) and P30CA46592 (Rogel Cancer Center).
Supplementary Material
References
- 1.Mizrahi J.D., Surana R., Valle J.W., Shroff R.T. Pancreatic cancer. Lancet. 2020;395:2008–2020. doi: 10.1016/S0140-6736(20)30974-0. [DOI] [PubMed] [Google Scholar]
- 2.Lippi G., Mattiuzzi C. The global burden of pancreatic cancer. Arch Med Sci. 2020;16:820–824. doi: 10.5114/aoms.2020.94845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vakoc C.R., Tuveson D.A. Untangling the genetics from the epigenetics in pancreatic cancer metastasis. Nat Genet. 2017;49:323–324. doi: 10.1038/ng.3798. [DOI] [PubMed] [Google Scholar]
- 4.Makohon-Moore A.P., Zhang M., Reiter J.G., Bozic I., Allen B., Kundu D., Chatterjee K., Wong F., Jiao Y., Kohutek Z.A., Hong J., Attiyeh M., Javier B., Wood L.D., Hruban R.H., Nowak M.A., Papadopoulos N., Kinzler K.W., Vogelstein B., Iacobuzio-Donahue C.A. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat Genet. 2017;49:358–366. doi: 10.1038/ng.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yachida S., Jones S., Bozic I., Antal T., Leary R., Fu B., Kamiyama M., Hruban R.H., Eshleman J.R., Nowak M.A., Velculescu V.E., Kinzler K.W., Vogelstein B., Iacobuzio-Donahue C.A. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roe J.S., Hwang C.I., Somerville T.D.D., Milazzo J.P., Lee E.J., Da Silva B., Maiorino L., Tiriac H., Young C.M., Miyabayashi K., Filippini D., Creighton B., Burkhart R.A., Buscaglia J.M., Kim E.J., Grem J.L., Lazenby A.J., Grunkemeyer J.A., Hollingsworth M.A., Grandgenett P.M., Egeblad M., Park Y., Tuveson D.A., Vakoc C.R. Enhancer reprogramming promotes pancreatic cancer metastasis. Cell. 2017;170:875–88 e20. doi: 10.1016/j.cell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waddell N., Pajic M., Patch A.M., Chang D.K., Kassahn K.S., Bailey P., Johns A.L., Miller D., Nones K., Quek K., Quinn M.C., Robertson A.J., Fadlullah M.Z., Bruxner T.J., Christ A.N., Harliwong I., Idrisoglu S., Manning S., Nourse C., Nourbakhsh E., Wani S., Wilson P.J., Markham E., Cloonan N., Anderson M.J., Fink J.L., Holmes O., Kazakoff S.H., Leonard C., Newell F., Poudel B., Song S., Taylor D., Waddell N., Wood S., Xu Q., Wu J., Pinese M., Cowley M.J., Lee H.C., Jones M.D., Nagrial A.M., Humphris J., Chantrill L.A., Chin V., Steinmann A.M., Mawson A., Humphrey E.S., Colvin E.K., Chou A., Scarlett C.J., Pinho A.V., Giry-Laterriere M., Rooman I., Samra J.S., Kench J.G., Pettitt J.A., Merrett N.D., Toon C., Epari K., Nguyen N.Q., Barbour A., Zeps N., Jamieson N.B., Graham J.S., Niclou S.P., Bjerkvig R., Grutzmann R., Aust D., Hruban R.H., Maitra A., Iacobuzio-Donahue C.A., Wolfgang C.L., Morgan R.A., Lawlor R.T., Corbo V., Bassi C., Falconi M., Zamboni G., Tortora G., Tempero M.A., Australian Pancreatic Cancer Genome I. Gill A.J., Eshleman J.R., Pilarsky C., Scarpa A., Musgrove E.A., Pearson J.V., Biankin A.V., Grimmond S.M. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailey P., Chang D.K., Nones K., Johns A.L., Patch A.M., Gingras M.C., Miller D.K., Christ A.N., Bruxner T.J., Quinn M.C., Nourse C., Murtaugh L.C., Harliwong I., Idrisoglu S., Manning S., Nourbakhsh E., Wani S., Fink L., Holmes O., Chin V., Anderson M.J., Kazakoff S., Leonard C., Newell F., Waddell N., Wood S., Xu Q., Wilson P.J., Cloonan N., Kassahn K.S., Taylor D., Quek K., Robertson A., Pantano L., Mincarelli L., Sanchez L.N., Evers L., Wu J., Pinese M., Cowley M.J., Jones M.D., Colvin E.K., Nagrial A.M., Humphrey E.S., Chantrill L.A., Mawson A., Humphris J., Chou A., Pajic M., Scarlett C.J., Pinho A.V., Giry-Laterriere M., Rooman I., Samra J.S., Kench J.G., Lovell J.A., Merrett N.D., Toon C.W., Epari K., Nguyen N.Q., Barbour A., Zeps N., Moran-Jones K., Jamieson N.B., Graham J.S., Duthie F., Oien K., Hair J., Grutzmann R., Maitra A., Iacobuzio-Donahue C.A., Wolfgang C.L., Morgan R.A., Lawlor R.T., Corbo V., Bassi C., Rusev B., Capelli P., Salvia R., Tortora G., Mukhopadhyay D., Petersen G.M., Australian Pancreatic Cancer Genome I., Munzy D.M., Fisher W.E., Karim S.A., Eshleman J.R., Hruban R.H., Pilarsky C., Morton J.P., Sansom O.J., Scarpa A., Musgrove E.A., Bailey U.M., Hofmann O., Sutherland R.L., Wheeler D.A., Gill A.J., Gibbs R.A., Pearson J.V., Waddell N., Biankin A.V., Grimmond S.M. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 9.Schulz W.A., Lang A., Koch J., Greife A. The histone demethylase UTX/KDM6A in cancer: progress and puzzles. Int J Cancer. 2019;145:614–620. doi: 10.1002/ijc.32116. [DOI] [PubMed] [Google Scholar]
- 10.Ler L.D., Ghosh S., Chai X., Thike A.A., Heng H.L., Siew E.Y., Dey S., Koh L.K., Lim J.Q., Lim W.K., Myint S.S., Loh J.L., Ong P., Sam X.X., Huang D., Lim T., Tan P.H., Nagarajan S., Cheng C.W., Ho H., Ng L.G., Yuen J., Lin P.H., Chuang C.K., Chang Y.H., Weng W.H., Rozen S.G., Tan P., Creasy C.L., Pang S.T., McCabe M.T., Poon S.L., Teh B.T. Loss of tumor suppressor KDM6A amplifies PRC2-regulated transcriptional repression in bladder cancer and can be targeted through inhibition of EZH2. Sci Transl Med. 2017;9:eaai8312. doi: 10.1126/scitranslmed.aai8312. [DOI] [PubMed] [Google Scholar]
- 11.Stief S.M., Hanneforth A.L., Weser S., Mattes R., Carlet M., Liu W.H., Bartoschek M.D., Dominguez Moreno H., Oettle M., Kempf J., Vick B., Ksienzyk B., Tizazu B., Rothenberg-Thurley M., Quentmeier H., Hiddemann W., Vosberg S., Greif P.A., Metzeler K.H., Schotta G., Bultmann S., Jeremias I., Leonhardt H., Spiekermann K. Loss of KDM6A confers drug resistance in acute myeloid leukemia. Leukemia. 2020;34:50–62. doi: 10.1038/s41375-019-0497-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taube J.H., Sphyris N., Johnson K.S., Reisenauer K.N., Nesbit T.A., Joseph R., Vijay G.V., Sarkar T.R., Bhangre N.A., Song J.J., Chang J.T., Lee M.G., Soundararajan R., Mani S.A. The H3K27me3-demethylase KDM6A is suppressed in breast cancer stem-like cells, and enables the resolution of bivalency during the mesenchymal-epithelial transition. Oncotarget. 2017;8:65548–65565. doi: 10.18632/oncotarget.19214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soto D.R., Barton C., Munger K., McLaughlin-Drubin M.E. KDM6A addiction of cervical carcinoma cell lines is triggered by E7 and mediated by p21CIP1 suppression of replication stress. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu W., Huang W., Yang Y., Qiu R., Zeng Y., Hou Y., Sun G., Shi H., Leng S., Feng D., Chen Y., Wang S., Teng X., Yu H., Wang Y. GATA3 recruits UTX for gene transcriptional activation to suppress metastasis of breast cancer. Cell Death Dis. 2019;10:832. doi: 10.1038/s41419-019-2062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andricovich J., Perkail S., Kai Y., Casasanta N., Peng W., Tzatsos A. Loss of KDM6A activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to BET inhibitors. Cancer Cell. 2018;33:512–526 e8. doi: 10.1016/j.ccell.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe S., Shimada S., Akiyama Y., Ishikawa Y., Ogura T., Ogawa K., Ono H., Mitsunori Y., Ban D., Kudo A., Yamaoka S., Tanabe M., Tanaka S. Loss of KDM6A characterizes a poor prognostic subtype of human pancreatic cancer and potentiates HDAC inhibitor lethality. Int J Cancer. 2019;145:192–205. doi: 10.1002/ijc.32072. [DOI] [PubMed] [Google Scholar]
- 17.Paulsen M.T., Veloso A., Prasad J., Bedi K., Ljungman E.A., Magnuson B., Wilson T.E., Ljungman M. Use of Bru-Seq and BruChase-Seq for genome-wide assessment of the synthesis and stability of RNA. Methods. 2014;67:45–54. doi: 10.1016/j.ymeth.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loomans H.A., Andl C.D. Intertwining of activin A and TGFβ signaling: dual roles in cancer progression and cancer cell invasion. Cancers (Basel) 2014;7:70–91. doi: 10.3390/cancers7010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Togashi Y., Kogita A., Sakamoto H., Hayashi H., Terashima M., de Velasco M.A., Sakai K., Fujita Y., Tomida S., Kitano M., Okuno K., Kudo M., Nishio K. Activin signal promotes cancer progression and is involved in cachexia in a subset of pancreatic cancer. Cancer Lett. 2015;356:819–827. doi: 10.1016/j.canlet.2014.10.037. [DOI] [PubMed] [Google Scholar]
- 20.Namwanje M., Brown C.W. Activins and inhibins: roles in development, physiology, and disease. Cold Spring Harb Perspect Biol. 2016;8:a021881. doi: 10.1101/cshperspect.a021881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawaguchi Y., Cooper B., Gannon M., Ray M., MacDonald R.J., Wright C.V. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- 22.Tuveson D.A., Shaw A.T., Willis N.A., Silver D.P., Jackson E.L., Chang S., Mercer K.L., Grochow R., Hock H., Crowley D., Hingorani S.R., Zaks T., King C., Jacobetz M.A., Wang L., Bronson R.T., Orkin S.H., DePinho R.A., Jacks T. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 23.Hingorani S.R., Petricoin E.F., Maitra A., Rajapakse V., King C., Jacobetz M.A., Ross S., Conrads T.P., Veenstra T.D., Hitt B.A., Kawaguchi Y., Johann D., Liotta L.A., Crawford H.C., Putt M.E., Jacks T., Wright C.V., Hruban R.H., Lowy A.M., Tuveson D.A. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 24.Hingorani S.R., Wang L., Multani A.S., Combs C., Deramaudt T.B., Hruban R.H., Rustgi A.K., Chang S., Tuveson D.A. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 25.Yadav D., Lowenfels A.B. The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology. 2013;144:1252–1261. doi: 10.1053/j.gastro.2013.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guerra C., Schuhmacher A.J., Cañamero M., Grippo P.J., Verdaguer L., Pérez-Gallego L., Dubus P., Sandgren E.P., Barbacid M. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Azizi N., Toma J., Martin M., et al. Loss of activating transcription factor 3 prevents KRAS-mediated pancreatic cancer. Oncogene. 2021;40(17):3118–3135. doi: 10.1038/s41388-021-01771-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antsiferova M., Werner S. The bright and the dark sides of activin in wound healing and cancer. J Cell Sci. 2012;125:3929–3937. doi: 10.1242/jcs.094789. [DOI] [PubMed] [Google Scholar]
- 30.Wiater E., Vale W. Roles of activin family in pancreatic development and homeostasis. Mol Cell Endocrinol. 2012;359:23–29. doi: 10.1016/j.mce.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 31.Zhao Y., Wu Z., Chanal M., Guillaumond F., Goehrig D., Bachy S., Principe M., Ziverec A., Flaman J.M., Collin G., Tomasini R., Pasternack A., Ritvos O., Vasseur S., Bernard D., Hennino A., Bertolino P. Oncogene-induced senescence limits the progression of pancreatic neoplasia through production of activin A. Cancer Res. 2020;80:3359–3371. doi: 10.1158/0008-5472.CAN-19-3763. [DOI] [PubMed] [Google Scholar]
- 32.Qiu W, Tang SM, Lee S, et al. Loss of activin receptor type 1B accelerates development of intraductal papillary mucinous neoplasms in mice with activated KRAS. Gastroenterology. 2016;150(1):218–228. doi: 10.1053/j.gastro.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding H., Zhang G., Sin K.W., Liu Z., Lin R.K., Li M., Li Y.P. Activin A induces skeletal muscle catabolism via p38β mitogen-activated protein kinase. J Cachexia Sarcopenia Muscle. 2017;8:202–212. doi: 10.1002/jcsm.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paajanen J., Ilonen I., Lauri H., Jarvinen T., Sutinen E., Ollila H., Rouvinen E., Lemstrom K., Rasanen J., Ritvos O., Koli K., Myllarniemi M. Elevated circulating activin A levels in patients with malignant pleural mesothelioma are related to cancer cachexia and reduced response to platinum-based chemotherapy. Clin Lung Cancer. 2020;21:e142–e150. doi: 10.1016/j.cllc.2019.10.013. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen N., Bellile E., Thomas D., McHugh J., Rozek L., Virani S., Peterson L., Carey T.E., Walline H., Moyer J., Spector M., Perim D., Prince M., McLean S., Bradford C.R., Taylor J.M., Wolf G.T. Tumor infiltrating lymphocytes and survival in patients with head and neck squamous cell carcinoma. Head Neck. 2016;38:1074–1084. doi: 10.1002/hed.24406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paulsen M.T., Veloso A., Prasad J., Bedi K., Ljungman E.A., Tsan Y.C., Chang C.W., Tarrier B., Washburn J.G., Lyons R., Robinson D.R., Kumar-Sinha C., Wilson T.E., Ljungman M. Coordinated regulation of synthesis and stability of RNA during the acute TNF-induced proinflammatory response. Proc Natl Acad Sci U S A. 2013;110:2240–2245. doi: 10.1073/pnas.1219192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bedi K., Paulsen M.T., Wilson T.E., Ljungman M. Characterization of novel primary miRNA transcription units in human cells using Bru-seq nascent RNA sequencing. NAR Genom Bioinform. 2020;2:lqz014. doi: 10.1093/nargab/lqz014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirkconnell K.S., Magnuson B., Paulsen M.T., Lu B., Bedi K., Ljungman M. Gene length as a biological timer to establish temporal transcriptional regulation. Cell Cycle. 2017;16:259–270. doi: 10.1080/15384101.2016.1234550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perusina Lanfranca M., Zhang Y., Girgis A., Kasselman S., Lazarus J., Kryczek I., Delrosario L., Rhim A., Koneva L., Sartor M., Sun L., Halbrook C., Nathan H., Shi J., Crawford H.C., Pasca di Magliano M., Zou W., Frankel T.L. Interleukin 22 signaling regulates acinar cell plasticity to promote pancreatic tumor development in mice. Gastroenterology. 2020;158:1417–1432.e11. doi: 10.1053/j.gastro.2019.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ettel M., Zhao L., Schechter S., Shi J. Expression and prognostic value of NSD1 and SETD2 in pancreatic ductal adenocarcinoma and its precursor lesions. Pathology. 2019;51:392–398. doi: 10.1016/j.pathol.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crowe A.R., Yue W. Semi-quantitative determination of protein expression using immunohistochemistry staining and analysis: an integrated protocol. Bio Protoc. 2019;9:e3465. doi: 10.21769/BioProtoc.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young C.C., Baker R.M., Howlett C.J., Hryciw T., Herman J.E., Higgs D., Gibbons R., Crawford H., Brown A., Pin C.L. The loss of ATRX increases susceptibility to pancreatic injury and oncogenic KRAS in female but not male mice. Cell Mol Gastroenterol Hepatol. 2019;7:93–113. doi: 10.1016/j.jcmgh.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang C., Lee J.E., Cho Y.W., Xiao Y., Jin Q., Liu C., Ge K. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci U S A. 2012;109:15324–15329. doi: 10.1073/pnas.1204166109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackson E., Willis N., Mercer K., Bronson R., Crowley D., Montoya R., Jacks T., Tuveson D. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olive K., Tuveson D., Ruhe Z., Yin B., Willis N., Bronson R., Crowley D., Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.