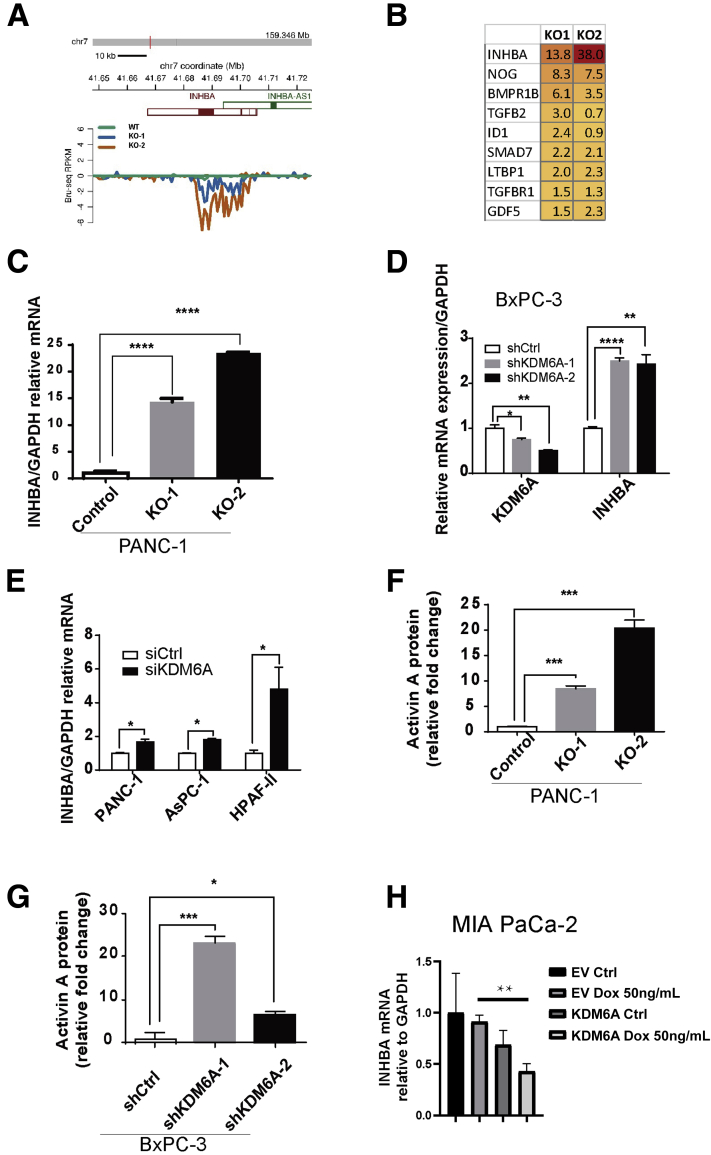
Figure 6.
KDM6A deficiency up-regulates activin A expression. (A) Reads per kilobase of transcript per million mapped reads (RPKM) of INHBA transcripts in KDM6A-KO and wild-type PANC-1 cells by Bru-seq analysis. (B) A heat map showing a list of genes in the TGF-β pathway that were up-regulated more than 1.5-fold in at least 1 clone of KDM6A-KO PANC-1 cells compared with control cells based on the Bru-seq data. (C) Real-time reverse-transcription polymerase chain reaction of INHBA in control and KDM6A-KO PANC-1 cells, (D) control and KDM6A short hairpin (sh) RNA knockdown BxPC-3 cells, and (E) control and KDM6A small interfering (si) RNA knockdown PANC-1, AsPC-1, and HPAF-II PDAC cell lines. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the reference gene. (F) Activin A protein levels in conditioned media from cultured control and KDM6A-KO PANC-1 cells and (G) shRNA control and KDM6A knockdown BxPC-3 cells measured by the enzyme-linked immunosorbent assay. (H) Real-time reverse-transcription polymerase chain reaction of INHBA in empty vector (EV) or a KDM6A inducible construct (KDM6A) transfected MIA PaCa-2 cells treated with vehicle control (Ctrl) or Doxycycline (Dox 50 ng/mL) to induce KDM6A expression. GAPDH was used as the reference gene. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001; unpaired t test. chr, chromosome.