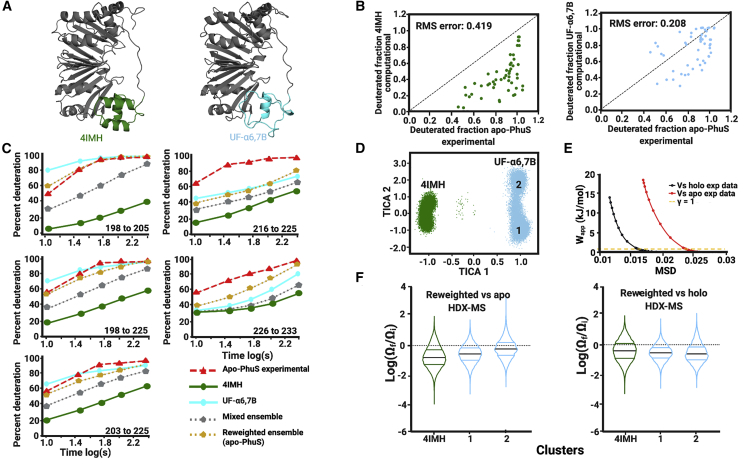
Figure 2.
Modeled local unfolding of α6 and α7 improves correlation to apo-PhuS HDX-MS data. (A) The crystal structure of apo-PhuS (PDB: 4IMH) and the best fitting model UF-α6,7B, with the α6,7,8 region highlighted (green and cyan, respectively). (B) Plots of the calculated deuterium uptake versus experimentally measured apo-PhuS deuterium uptake for all peptides spanning the α6,7,8 motif and deuterium exchange reaction time points up to 5 min. The calculated deuterium uptake was obtained from MD simulations of the apo-PhuS structure 4IMH (left) or of the UF-α6,7B locally unfolded model (right); the RMS error to the line of identity is also shown. (C) Kinetic deuterium uptake plots of representative peptides in the α6,7,8 region including the traces for the apo-PhuS experimental data (red dashed line), the 4IMH MD simulations (green solid line), the UF-α6,7B MD simulations (cyan solid line), the mixed ensemble (silver dotted line), and the HDXer reweighted ensemble (gold dotted line). (D) TICA separation of the mixed ensemble. Frames are mapped based on simulation of origin (4IMH, green and UF-α6,7B, cyan). (E) The work versus MSD plots or decision plot obtained for the reweighting the mixed ensemble to the experimental apo-PhuS target data (red) and to the experimental holo-PhuS target data (black) with the chosen γ-value of 1 displayed as the dashed yellow line. (F) Violin plots of the log(Ωf/Ωi) of individual frames mapped to the clusters generated by TICA and colored accordingly. Within the violins, black lines indicate the average log(Ωf/Ωi) and the color lines bracket 50% of the population.