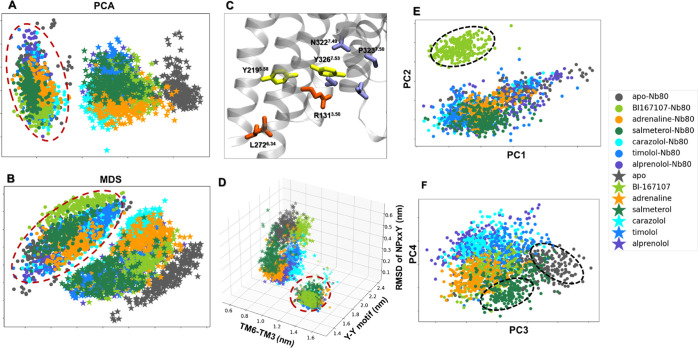
Figure 2.
Dimensionality reduction analysis applied to the active-like simulation ensembles. Each point represents a snapshot and is depicted according to the ligand and Nb80-bound ensembles. Input features are the residue–residue Cα atom distances. The Nb80-bound ensembles are highlighted by a red dashed circle. (A) Principal component analysis (PCA) and (B) Multidimensional scaling (MDS) projection on the first two principal components extracted from all trajectories (combining trajectories with and without Nb80). (C) Conserved microswitches of the β2AR: R131 and L272 (orange) are located in the transmembrane 3 (TM3) and TM6, respectively. The outward displacement of TM6 is represented by the distance between the Cα atoms of R1313.50 and L2726.34. Y2195.58 and Y3267.53 (yellow) are part of TM5 and TM7, which are close to each other via a water-mediated interaction (Y-Y motif) in the β2AR active state. The N7.49P7.50xxY7.53 motif (blue) is at the bottom of TM7. (D) Distributions of the distances between TM6 and TM3, and Y-Y and RMSD of NPxxY motifs. (E, F) PCA projection onto the first four principal components (PC) of the Nb80-bound trajectories only. BI167107-, salmeterol-bound, and apo snapshots are highlighted by black dashed lines.